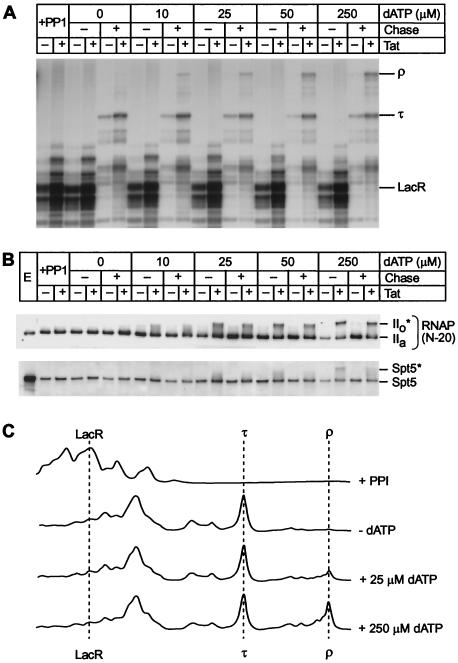
FIG. 8.
Activation of elongation complexes by phosphorylation prior to elongation. (A) Transcription. Elongation complexes were assembled by using the pW1 DNA template in the presence of lac repressor and dephosphorylated (+PP1) as described in the legend to Fig. 6. The complexes were rephosphorylated by adding between 0 and 250 μM dATP as a phosphate donor. After a brief wash to remove unincorporated dATP, 100 μM DRB was then added to prevent further phosphorylation of the complex during the chase reaction. The rephosphorylated complexes were chased by addition of all four ribonucleotide triphosphates and 25 mM IPTG. Positions of transcripts at the runoff (ρ), terminator (τ), and lac repressor (LacR) are shown. (B) Immunoblot. Samples from the reactions shown in panel A were immunoblotted with N-20 antibody against RNA Pol II and the anti-Spt5 antibody. (C) Quantitative analysis of the gel shown in panel A by densitometry. Note that there is a strong correlation between the extent of CTD phosphorylation and the amount of the runoff product (ρ).