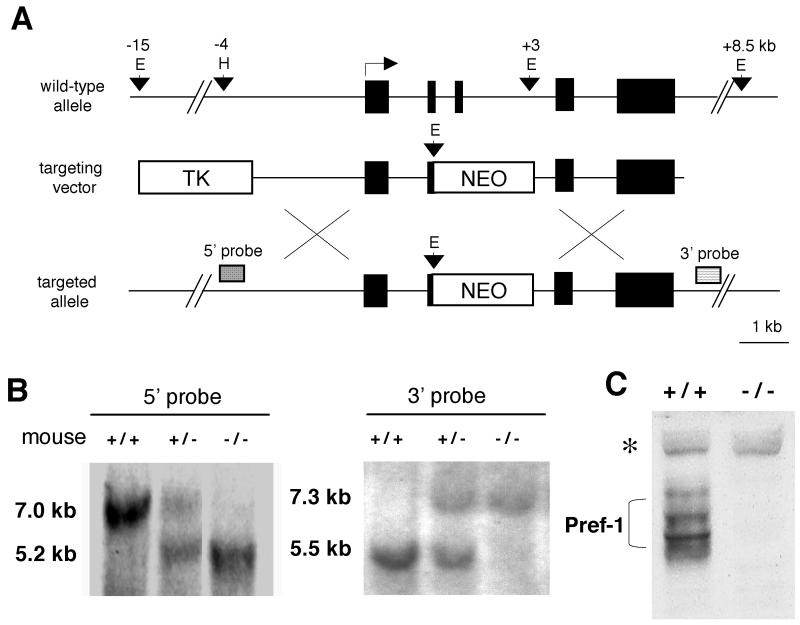
FIG. 1.
Targeted disruption of the pref-1 gene. (A) The top diagram is a schematic representation of the wild-type pref-1 allele with five exons. Pref-1 exons are represented by black boxes. The middle diagram shows the targeting construct with pPNT-NEO and thymidine kinase (TK) as positive and negative selectable markers, respectively. The bottom diagram shows the pref-1 allele mutated by homologous recombination. The probes used for Southern blot analyses and the sizes of the restriction fragments detected are indicated. E, EcoRI; H, HindIII; X, XbaI. (B) Southern blot analysis of EcoRI-cut (3′ probe) and EcoRI-HindIII-cut (5′ probe) mouse tail DNA prepared from F2 mice generated by intermating F1 heterozygotes (pref-1+/−). The 7.0- and 5.5-kb bands represent the wild-type allele, and the 5.2- and 7.3-kb bands represent pref-1-null alleles. (C) Western blot analysis of Pref-1 in embryos (E17.5). A polyclonal antibody against mouse Pref-1 was used. The embryos were generated from crossbreeding pref-1 heterozygotes. The multiple forms of Pref-1 produced by alternative splicing and posttranslational modification (bracketed) are detected in wild-type but not in homozygous (pref-1−/−) embryos. ∗, nonspecific band indicating equal amounts of sample loading.