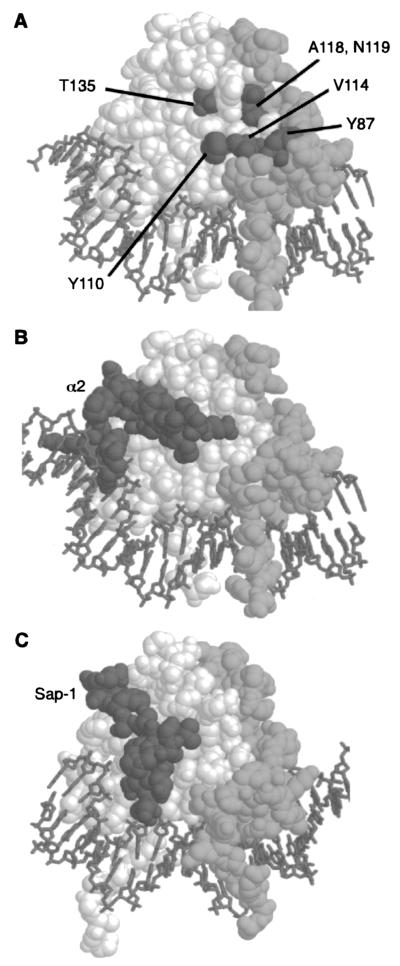
FIG. 7.
Position of the residues required for Arg80 activity. (A) The relative positions of residues that affect Arg80 function are displayed on a model of the Mcm1 MADS box domain dimer (spacefill) bound to DNA (stick-like figures). One monomer is shown in white, and the other is shown in light gray. Residues corresponding to positions in Arg80 that are required for regulation of arginine metabolism are shown in dark gray. These residues cluster in a partially contiguous face on the side of the protein that is part of the dimer interface. Residue Y87 is from one monomer, while residues Y110, V114, A118, N119, and T135 are from the other monomer. (B) Contacts between α2 and Mcm1 are in a different region of the MADS box domain. A complex of the α2 linker (dark gray) binding to the Mcm1 dimer is shown. The Mcm1 dimer is in roughly the same orientation as that shown in panel A. (C) Contacts between the SAP-1 and SRF are in a different region of the MADS box domain. A complex of the SAP-1 B-box (dark gray) binding to the SRF dimer is shown. The SRF dimer is in roughly the same orientation as that of the Mcm1 dimer shown in panel A.