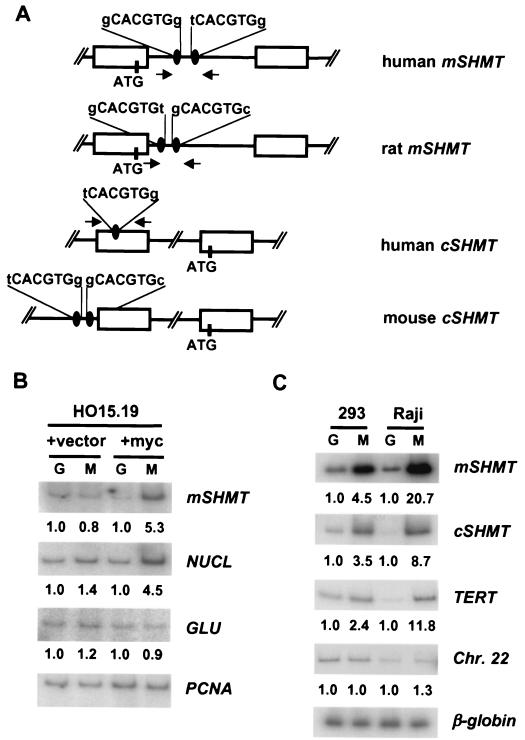
FIG. 4.
c-Myc binds to the promoters of mSHMT and cSHMT genes in vivo. (A) Schematic presentation of human, mouse, and rat mSHMT and cSHMT promoters. Open boxes represent exons. Myc binding sites and ATG codons are shown. Arrows indicate the positions and direction of primers used in PCR. (B and C) HO15.19 cells infected with empty vector (+vector lanes), HO15.19 cells infected with c-myc-expression vector (+myc lanes), HEK 293 cells, or Raji cells were cross-linked and lysed, and chromatin was immunoprecipitated with c-Myc-specific (M lanes) or Gal4-specific (G lanes) antibodies, followed by the reversal of the cross-linking and DNA isolation. Isolated DNA was used in PCR with radiolabeled oligonucleotides flanking Myc binding sites in the promoters of the genes indicated to the right of the blots. For negative controls, two types of PCR were performed. The first one was carried out with oligonucleotides flanking CACGTG sites that do not interact with c-Myc in the promoter of the rat glucokinase gene (GLU) (11) (B) or somewhere on human chromosome 22 (Chr. 22) (4) (C). Another PCR was performed with oligonucleotides flanking a DNA region containing no CACGTG sites in the rat PCNA gene (B) or in the third intron of a silent human β-globin gene (C). Quantitation of the PCR products was performed with the Molecular Dynamics IhoshphorImaging system. A number below the a blot indicates the fold difference in the signal intensity, determined by dividing a given PCR signal by a corresponding PCNA-specific (B) or β-globin-specific (C) signal and by the ratio of these signals in the corresponding Gal4 lane.