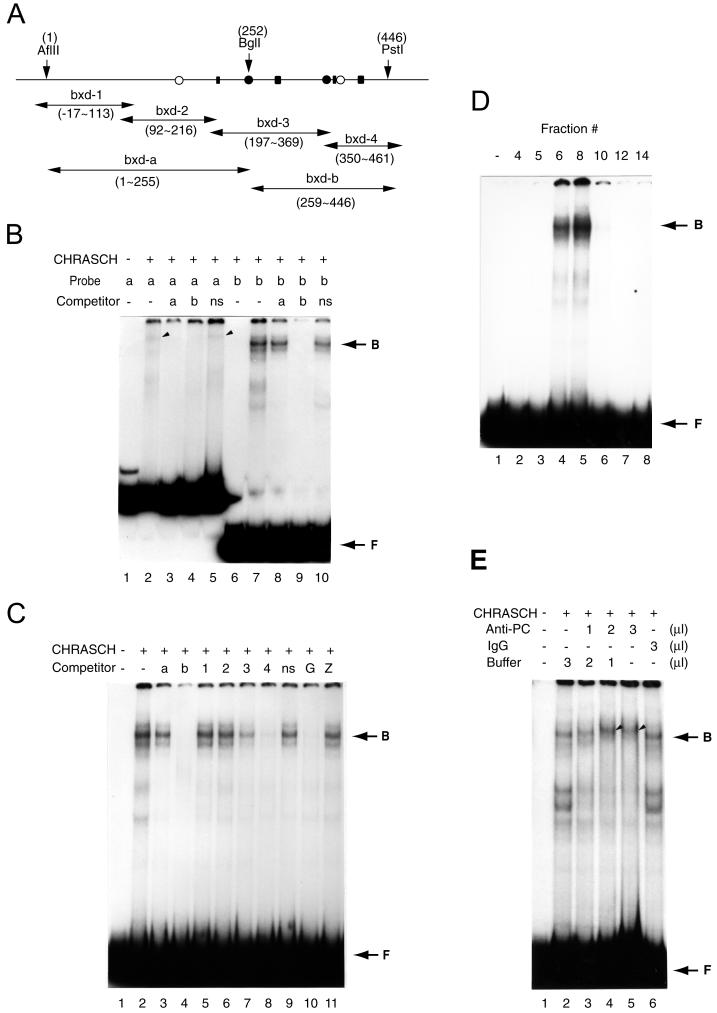
FIG. 2.
CHRASCH binds the (GA)n motif. (A) Map of DNA fragments in B-151 used for binding and competition assays. The locations of the (GA)n motif (solid rectangular box), YY1 (solid circles), and ZESTE (open circles) binding sites are shown. The coordinates of restriction sites and various fragments are indicated. (B) Preferential binding of CHRASCH to the bxd-b fragment. Binding of labeled bxd-a (a) or bxd-b (b) probe was carried out with or without (−) an excess amount of unlabeled bxd-a or bxd-b or a DNA fragment from the polylinker of a pBluescribe vector(ns). Specific binding to bxd-a and bxd-b probes is indicated by the arrowhead and arrow, respectively. (C) Binding of CHRASCH to the (GA)n motif. Binding assays were carried out with the bxd-b probe. The competitors were bxd-a (a), bxd-b (b), bxd-1 to -4 (1 to 4), linker DNA (ns), or fragments containing multiple (GA)n (G) or ZESTE (Z) binding sites. (D) Elution profile of the DNA binding activity. Aliquots (2 μl) of eluted fractions (same fractions as shown in Fig. 1A) were assayed with the bxd-b probe. (E) Antibody supershift assay. Affinity-purified PC antibody (2.5 mg/ml), IgG (2.5 mg/ml), or buffer were preincubated with CHRASCH before the addition of a reaction mixture containing the bxd-b probe. The volume of antibody solution is indicated. The supershifted complex is indicated by the arrowhead. Note that fast-migrating bands appeared after 1 h of preincubation (lanes 2 and 6), suggesting instability of the complex. The EMSA shown here was done with 3.5% polyacrylamide gels. B, bound DNA-protein complex; F, free probe.