Abstract
Telomerase is a ribonucleoprotein particle (RNP) involved in chromosome end replication, but its biogenesis is poorly understood. The RNA component of yeast telomerase (Tlc1) is synthesized as a polyadenylated precursor and then processed to a mature poly(A)− form. We report here that the karyopherin Mtr10p is required for the normal accumulation of mature Tlc1 and its proper localization to the nucleus. Neither TLC1 transcription nor the stability of poly(A)− Tlc1 is significantly affected in mtr10Δ cells. Tlc1 was mostly nuclear in a wild-type background, and this localization was not affected by mutations in other telomerase components. Strikingly, in the absence of Mtr10p, Tlc1 was found dispersed throughout the entire cell. Our results are compatible with two alternative models. First, Mtr10p may import a cytoplasmic complex containing Tlc1 and perhaps other components of telomerase, and shuttling of Tlc1 from the nucleus to the cytoplasm and back may be necessary for the biogenesis of telomerase (the “shuttling” model). Second, Mtr10p may be necessary for the nuclear import of some enzyme needed for the nuclear processing and maturation of Tlc1, and in the absence of this maturation, poly(A)+ Tlc1 is aberrantly exported to the cytoplasm (the “processing enzyme” model).
Telomeres, the ends of linear chromosomes, cannot be fully replicated by conventional DNA polymerases (21, 35, 53). This end replication problem is solved in most eukaryotes by a ribonucleoprotein complex called telomerase, which extends the natural 3′ ends of chromosomes by adding a repetitive sequence copied from its RNA component (reviewed in references 12 and 36). Telomerases from different organisms contain a conserved reverse transcriptase subunit, an RNA subunit that provides the template for the newly synthesized telomeric DNA, and also a set of additional proteins (showing little or no sequence conservation) serving a variety of accessory functions (reviewed in reference 33). Little is known, however, about the biogenesis of telomerase, i.e., the processing, transport, and assembly of its different components.
In the yeast Saccharomyces cerevisiae, the reverse transcriptase is called the Est2 protein (7, 22), and the template RNA is called Tlc1 (47). In addition, yeast telomerase contains at least two accessory proteins, named Est1 and Est3, which are required for telomerase-dependent telomere elongation in vivo (14, 18, 20, 49). Est1 binds Tlc1 and mediates the access of telomerase to the telomere (9, 51, 57), whereas the function of Est3 is still unknown. Yeast telomerase complex also contains Sm proteins (45), which are common components of the small nuclear ribonucleoprotein particles (snRNPs) (reviewed in reference 54). In fact, Tlc1 shares a number of structural and biogenetic hallmarks with the uridine-rich snRNAs (UsnRNAs) U1, U2, U4, and U5, which are subunits of the snRNPs. In addition to their high uridine content, snRNAs and Tlc1 are RNA polymerase II transcripts; they have a 5′-2,2,7-trimethylguanosine (TMG) cap; and they bind the Sm proteins through a specific sequence termed the Sm site, which is essential for their accumulation (4, 45; snRNP biogenesis has been reviewed in reference 55). In yeast, two distinct forms of Tlc1 RNA are found: a short-lived polyadenylated transcript [poly(A)+ Tlc1] and a much more abundant and stable poly(A)− species (mTlc1), which likely derives from the poly(A)+ form (4). Although no poly(A)+ fractions of snRNAs are detected under physiological conditions, poly(A)+ forms of these RNAs have been reported under conditions which interfere with the normal processing of snRNAs (8, 32, 43, 50), suggesting that the poly(A)− mature snRNAs may derive from poly(A)+ precursors. Because of these parallels, it is possible that both snRNPs and telomerase may follow similar biogenesis pathways.
In higher eukaryotes, UsnRNAs are exported to the cytoplasm where they bind the Sm core proteins. Sm binding triggers the hypermethylation of the cap and the trimming of extra nucleotides (fewer than 20) at the 3′ end, and it generates a bipartite nuclear localization signal (NLS) required for nuclear import of the snRNP (reviewed in references 30, 31, and 55). One part of this NLS consists of the TMG cap, specifically recognized by the import factor Snurportin-1, which in turn serves as an adapter between the TMG cap and the general import factor importin-β (13, 37). Another part of the NLS resides in the Sm core of the particle, but its exact molecular nature and the import factor that interacts with it remain elusive. In yeast, it is not known whether UsnRNAs transit through the cytoplasm. In this respect, yeast orthologs of vertebrate PHAX (the specific mediator of snRNA export) and Snurportin-1 have not been identified (13, 34), which reinforces a model wherein snRNP biogenesis in yeast would be exclusively nuclear.
Nucleocytoplasmic transport is mediated by soluble receptors that recognize structural features (NLS or nuclear export signal) in their substrates. In general, the transport apparatus is versatile, with individual receptors binding a variety of cargoes and vice versa (reviewed in references 6, 11, and 31). Most transport receptors belong to a large family of homologous proteins known as karyopherins. They share limited sequence identity (15 to 25%), contain an N-terminal Ran-GTP-binding motif, and bind some components of the nuclear pore complex (reviewed in reference 31). One of the members of this family is S. cerevisiae Mtr10p (10, 38, 44). Although first identified in a screen for mutants defective in mRNA export, mtr10 cells also present abnormalities in different aspects of ribosome biogenesis (15, 29, 48). The major cargo of Mtr10p is the mRNA-binding protein Npl3 (38, 44), a nucleocytoplasmic shuttling protein involved in mRNA export and pre-rRNA processing (17, 40). Many of the phenotypes of mtr10 mutants can be at least partly attributed to impairment of Npl3p function (for example, see reference 29).
Here, we have isolated mtr10 in a genetic screen for telomere maintenance mutants. mtr10 (but not npl3) cells exhibited a gradual telomere shortening that can be explained by the low levels of Tlc1 in these mutants. Kinetics studies revealed a defective intermediate step in telomerase biogenesis, whereas the stability of mTlc1 was unaffected. By fluorescence in situ hybridization (FISH), we detected Tlc1 mostly in the cell nucleus in wild-type cells. This localization was independent of other telomerase components. By contrast, Tlc1 was found dispersed throughout the entire cell in an mtr10 background. Our data fit two alternative models wherein Mtr10p would play a role as an import receptor either of a Tlc1-containing cytoplasmic complex (the “shuttling” model) or of a protein involved in the nuclear processing of telomerase RNP (the “processing enzyme” model).
MATERIALS AND METHODS
Strains, plasmids, and genetic manipulations.
Growth media and general genetic manipulations were as previously described (25). Strains carrying a MATa-inc or a MATα-inc allele were used in the screen. These were made by crossing GRY1078a or GRY1060α (kindly provided by J. Strathern), respectively, to W303 MATα or MATa (ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 ssd1-d [psi+], backcrossing twice again to W303, and selecting GRY mating type and W303 markers. DF5 background strains, both wild type and mtr10::HIS3, were a gift from G. Blobel and have been described elsewhere (38). RS453 strains were from E. Hurt (44). The W303 mtr10Δ strain was constructed from RS453 mtr10::HIS3 by PCR amplification of the mtr10::HIS3 cassette and subsequent transformation of a W303 diploid strain. After sporulation, substitution of the wild-type allele for the mtr10::HIS3 cassette was confirmed by PCR and cosegregation with the slow-growth phenotype. S288C strains were purchased from Research Genetics (Huntsville, Ala.) and are part of the Saccharomyces genome deletion project (www-sequence.stanford.edu/group/yeast_deletion_project), wherein each open reading frame has been substituted by a KanMX4 module. These include mtr10Δ, npl3Δ, and gbp2Δ strains. S288C hrb1Δ was constructed from a hrb1::HIS3 strain (P. Silver) following a PCR-replacement strategy. Double and triple mutants were obtained by crossing and sporulation. In each case, they were confirmed by PCR. W303 yku80::KAN strain was a gift from S. Jackson. Construction of the W303 tlc1::LEU2 strain has been previously described (49). W303 tlc1::LEU2 mtr10::HIS3 double mutants were obtained by crossing the corresponding single mutants. Strains est1::HIS3, est2::URA3, est3::LYS2, cdc13est and the quadruple mutant used in the FISH experiments were obtained by sporulation of a heterozygous diploid (kindly provided by V. Lundblad).
Plasmid pBST3 was made as follows: oligos 5′-ATC AGC GGA TCC GTG TGT GGG TGT GTG TGT GGG TGT GTG TGT GGG TGT GTG TGT TTC AGC TTT CCG CAA CAG TAT AAT TTT CAA TTC AAT TCA TC-3′ and 5′-ATC AGC GGA TCC GTG TGT GGG TGT GTG TGT GGG TGT GTG TGT GGG TGT GTG TGT TTC AGC TTT CCG CAA CAG TAT AAC TTT TTC TTT CCA ATT TT-3′ were used to PCR amplify the URA3 gene from a URA3-containing plasmid. The resulting Tel-HO-URA3-HO-Tel cassette (where Tel is a 41-nucleotide [nt]-long TG-rich telomeric sequence and HO represents the 24-nt HO endonuclease recognition site) was flanked by BamHI restriction sites and inserted into the BamHI site of plasmid pRS315 (46). Plasmid pBST1 contained the HO endonuclease under the GAL10 promoter and was made by inserting a 3.3-kb EcoRI/SalI fragment from pGAL-HO (a gift from I. Herskowitz) into pRS313 (46). Plasmid pRS314-MTR10 was made by PCR amplification of MTR10 using oligos 5′-GCA CAG GAT CCG CAA TTT ATT GGT GGC ACC-3′ and 5′-AAA AGC GGC CGC ACC AGA ATG TGT GAA GAG-3′, then cloned as a 3.5-kb BamHI/NotI fragment into pRS314. The plasmids used in the suppression experiment (see Fig. 2B) were constructed by cutting previous plasmids and inserting the following fragments into pRS426 (a 2μm-based plasmid): NotI/XhoI TLC1 fragment from pVL796, HindIII/XbaI EST1 fragment from pVL887, SacI/XhoI EST2 fragment from pRS315-EST2, EcoRI/SalI EST3 fragment from pVL1014, and XbaI/KpnI CDC13 fragment from pVL440. The same sites were used as cloning sites in pRS426 except for SpeI, which was used instead of XbaI. The pVL plasmids were a generous gift from V. Lundblad. Plasmid pRS315-EST2 was from J. Zhou and B. Futcher (unpublished data). Plasmid pRS314 carrying the mtr10-7 allele was kindly provided by E. Hurt (44). Plasmid pSD171 (CEN, TRP1) carrying TLC1 under the GAL1-10 promoter was kindly given by D. Gottschling. Tlc1 from this plasmid contains 5′ extra nucleotides and consequently runs slightly slower than the endogenous Tlc1 (4). For the experiment shown below in Fig. 5A, the ≈3-kb GAL-TLC1 construct was cut with SalI and cloned at the XhoI site of pRS316 (CEN, URA3). Plasmids pPS1331 (2μm; LEU2 HRB1) and pPS1152 (2μm; URA3 NPL3) were used for HRB1 and NPL3 overexpression experiments and were provided by P. Silver.
FIG. 2.
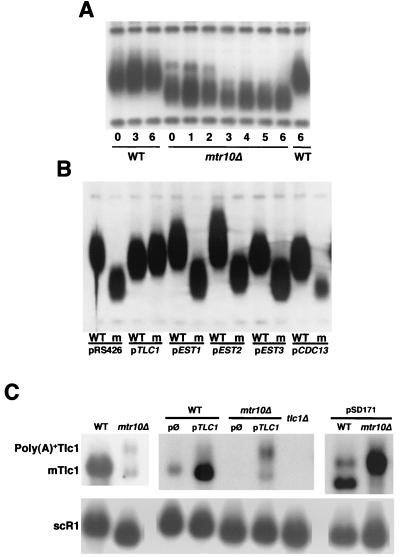
mtr10 cells are defective in the telomerase pathway. (A) Progressive telomere shortening in mtr10Δ mutants. A heterozygous diploid MTR10/mtr10Δ was sporulated and dissected. Spores germinated on YEPD plates for 4 days at 30°C. Wild-type (WT) and mutant strains were selected and then either inoculated in YEPD liquid and grown overnight for genomic DNA isolation (time 0) or streaked out on plates and grown for 24 h. This procedure was repeated for six consecutive days. The Southern blot shows only the Y′ class telomere band and size markers. Telomere length remained constant in a wild-type background throughout the course of the experiment but progressively diminished in the mutant. (B) Overexpression of TLC1 restored wild-type telomere length in the mtr10Δ mutant. Heterozygous diploid MTR10/mtr10Δ cells were transformed with the 2μm plasmid pRS426 or derivatives carrying genes TLC1, EST1, EST2, EST3, and CDC13. After sporulation, dissection, and germination, plasmid-bearing wild-type (WT) and mutant (m) strains were selected and streaked out for four successive days on medium lacking uracil. Again, only the Y′ class telomere band is shown. (C) Low levels of Tlc1 in mtr10Δ cells. Endogenous and overexpression levels of Tlc1, both poly(A)+ and mature (mTlc1) forms, were examined by Northern blotting. The level of endogenous mTlc1 was very low in an mtr10Δ mutant, whereas the poly(A)+ form was even easier to detect than in the wild-type strain. mTlc1 levels in mtr10Δ cells were greatly restored by overexpression of TLC1 from its own promoter by using a high-copy-number plasmid (pTLC1). By contrast, when overexpressed from the GAL promoter (pSD171), little, if any, mTlc1 accumulated in the mutant. Exposure time was different for the three panels presented. scR1 RNA is shown with identical exposure time. mtr10 mutations did not appreciably influence the levels of this RNA.
FIG. 5.
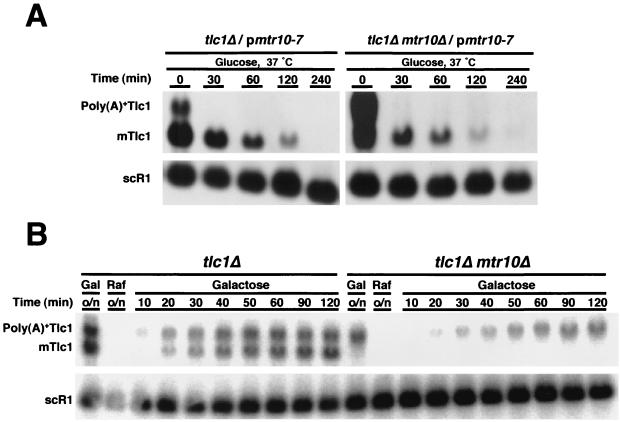
Defective biogenesis of Tlc1 in mtr10Δ strains. (A) The stability of mTlc1 did not depend on Mtr10 function. tlc1Δ MTR10 and tlc1Δ mtr10Δ strains carrying plasmids pmtr10-7 and pGAL-TLC1 were grown overnight at 18°C in galactose-containing medium. Cultures were shifted to 37°C for 2 h prior to addition of glucose. Samples were taken at the specified times, total RNA was extracted, and equal amounts of RNA were examined by Northern blot analysis. scR1 was probed as a loading reference. Signal intensities for Tlc1 were obtained with different exposures between strains. (B) mTlc1 fails to accumulate in mtr10Δ cells. Strains carrying pGAL-TLC1 were grown overnight (o/n) in raffinose-containing medium, then supplemented with galactose (2% final concentration), and sampled at the specified times. Equal amounts of total RNA from these samples were analyzed by Northern blotting. scR1 was used as a loading control. Samples from overnight-grown cells in galactose-containing medium are also shown (Gal).
Genetic screen.
MATa-inc and MATα-inc strains carrying plasmids pBST3 and pBST1 were mutagenized with ethyl methanesulfonate as previously described (56) and plated on medium lacking leucine and histidine. Colonies were replica plated on 5-fluoroorotic acid (5-FOA)- and galactose-containing medium with or without leucine. Cells unable to grow in the absence of leucine were picked up from the leucine-containing replica. These candidates were retested for their inability to maintain pBST3. They were crossed to W303 and sporulated, and their chromosomal telomere length was examined by Southern blot analysis. Those presenting a short telomere phenotype were backcrossed to wild-type W303 at least twice.
Cloning LPM25 and sequencing of the MTR10 locus in strain lpm25.
Strain lpm25 was transformed with a genomic yeast library cloned on a 2μm-based plasmid (YEp213) kindly given by S. Cameron and M. Wigler. This library was screened for complementation of the temperature-sensitive (Ts) phenotype of lpm25 cells. We carried out two independent transformations, examining about 25,000 transformants each time. Only one transformant in each case was able to grow at 37°C. These contained, respectively, a 7- and 14-kb overlapping genomic insert. Genes contained in both inserts were subcloned into pRS314 and tested independently for complementation. Only an MTR10-containing plasmid suppressed the phenotypes of lpm25 mutants.
Isolation of the MTR10 locus in strain lpm25 for further sequencing was carried out by a plasmid gap-repair strategy (24). Briefly, the library plasmid containing the 14-kb insert was cut with XhoI/PacI about 2,400 and 500 bp upstream and downstream of MTR10, respectively. The linear fragment was introduced into the lpm25 strain and colonies with unstable selectable marker (nonintegrated) were selected. Plasmids isolated from these colonies were unable to suppress the phenotypes of lpm25 cells. The MTR10 locus was sequenced from nt −275 to +1860. Since we found a nonsense mutation at position 1062 and PCR amplification suggested no major alterations (deletions or insertions), we did not sequence the gene further.
Southern and Northern blotting.
Genomic DNA was isolated with the Wizard genomic purification kit from Promega (Madison, Wis.), following the manufacturer's instructions. DNA was digested overnight with PstI, mixed with loading buffer and size markers (511 and 1,436 bp), and loaded on a 1% agarose gel. The blotting procedure was as previously described (3). Detection of telomeric bands was accomplished by using a 28-nt TG-rich oligo probe labeled with either 32P (Amersham Pharmacia, Piscataway, N.J.) or digoxigenin (Roche Molecular Biochemicals, Indianapolis, Ind.) and exposing the membrane to a Molecular Dynamics PhosphorImager or X-ray film, respectively.
Total RNA was isolated by a hot acidic phenol protocol (5). RNA was quantitated by fluorometry or spectrophotometry, and roughly equal amounts of total RNA (≈20 μg) were loaded onto formaldehyde-agarose gels as described in reference 27. Tlc1, U2, and ACT1 probes were labeled by random-primed DNA labeling (Roche Molecular Biochemicals) from restriction fragments containing the corresponding genes. Other probes to detect more abundant RNAs consisted of end-labeled oligos. 32P or digoxigenin was used.
FISH.
Cells were grown to an optical density at 600 nm (OD600) of 0.4 to 0.6, mixed with 37% formaldehyde (120 μl/ml of cells), and incubated for 1 h at room temperature (RT). After fixation, they were washed in PB (0.1 M KH2PO4, pH 6.5) and resuspended in 250 μl of PB with 1.2 M sorbitol. Cells were spheroplasted by treatment with 2 μl of Zymolyase 100T (10 mg/ml) for 30 min at 37°C, washed in PB plus 1.2 M sorbitol, and placed onto polylysine-treated slides. After washing the cells with PB, they were permeabilized with 0.1% Triton X-100 and then prehybridized with FHB solution (50% deionized formamide, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] [pH 8.0], 500 μg of yeast tRNA/ml, 5 mM EDTA [pH 8.0], 100 μg of heparin/ml, 0.05% Sarkosyl) for at least 1 h at RT in a humid chamber. Antisense RNA probes were labeled with Alexa Fluor 488 (Molecular Probes, Eugene, Oreg.) according to the manufacturer's instructions and hybridized overnight at 37°C in FHB. After hybridization, slides were washed twice in 0.2× SSC for 30 min at RT and once at 55°C. Cells were then washed with ABS4 (0.1 M Tris-HCl [pH 8.0], 125 mM NaCl, 10 mM MgCl2) and incubated with 5 μg of 4′,6′-diamidino-2-phenylindole (DAPI)/ml in ABS4 for DNA staining. Cells were washed again with ABS4, dried 10 min at RT in the dark, and mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, Calif.). Signal was visualized in a fluorescence microscope.
RESULTS
Identification of mtr10 as a telomere maintenance mutant.
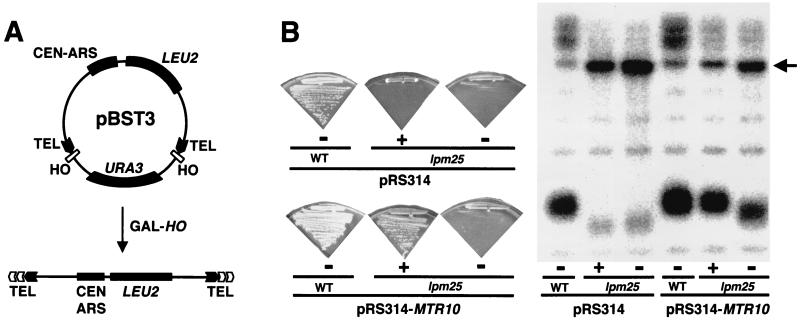
To identify novel mutants defective in telomere maintenance, we have designed a genetic screen based on a previous scheme (23). We constructed the LEU+ URA+ plasmid pBST3 (Fig. 1A). When pBST3 is cut with the HO endonuclease, it releases a short, linear, LEU+ ura− minichromosome whose ends are capped with 41 bp of telomeric sequence. In a wild-type strain, these 41-bp proto-telomeres nucleate the formation of full-length telomeres (26) and so allow the appearance of LEU+ ura− cells. However, in mutant strains defective for various steps in telomere addition or maintenance, these 41-bp proto-telomeres are not sufficient for maintenance of the linear plasmid and so no LEU+ ura− cells can be derived from such mutants.
FIG. 1.
mtr10 mutants have short telomeres. (A) Plasmid pBST3. Two cassettes, each consisting of an HO site followed by a 41-bp-long yeast telomeric sequence (TEL), flanked a copy of the URA3 gene as shown. Induction of the HO endonuclease releases a linear plasmid containing a LEU2 selectable marker. This plasmid can be efficiently maintained only when the exposed telomeric tracts are elongated to create a functional telomere. (B) MTR10 suppressed both the growth defect and the short telomere phenotype of lpm25 mutants. MTR10 was subcloned into pRS314 and transformed into lpm25 cells. Mutants carrying the MTR10 gene were indistinguishable from a wild-type strain. Vector alone did not produce any effect. Loss of MTR10 plasmid from lpm25 strains reverted both phenotypes. Plates were incubated at 30°C for 24 h. Some colonies from these plates were inoculated in liquid medium and grown to an OD600 of 1. Genomic DNA from these cultures was used in the Southern blotting. The arrow indicates a band corresponding to Y′ subtelomeric DNA. +, plasmid present; −, plasmid absent.
By selecting for the LEU2 marker and counterselecting against the URA3 marker using 5-FOA (1), we isolated 58 candidate mutants that failed to maintain linearized pBST3. We named these lpm mutants, for linear plasmid maintenance. Six of the 58 mutant isolates had abnormally short chromosomal telomeres, as assayed by Southern blotting (data not shown). Of these six, complementation and cosegregation analyses showed that two mutant isolates were alleles of EST2, the catalytic subunit of telomerase (7, 22), demonstrating that our screen was capable of detecting genes involved in telomere function. However, we did not detect alleles of EST1, EST3, or TLC1, indicating that the screen was not saturated.
Mutant lpm25 had short telomeres, poor growth at 30°C, and failed to grow at 37°C (Fig. 1B and data not shown). The growth phenotypes and the telomere shortening phenotype were tightly linked, suggesting they might be due to the same mutation. We cloned the wild-type LPM25 gene by complementation of the Ts lethal phenotype. Two independent complementing plasmids were analyzed; these had overlapping genomic inserts, which included the MTR10 gene. Subcloning of DNA in the overlapping region demonstrated that a portion of the insert containing only the MTR10 gene was capable of suppressing both the Ts growth defect and the telomere shortening in lpm25 cells (Fig. 1B and data not shown). These effects were due to MTR10, since lpm25 mutants that had lost the MTR10 plasmid or carried only empty vector displayed lpm25 mutant phenotypes (Fig. 1B). Interestingly, although lpm25 cells exhibited the full growth defect very rapidly after losing the MTR10 plasmid, telomeres in these cells shortened only gradually.
We next examined telomere length in four different genetic backgrounds (DF5, RS453, S288C, and W303) deleted for MTR10. All four mtr10Δ strains had shorter telomeres than their wild-type counterparts (data not shown). Sequencing of the MTR10 locus in the lpm25 strain revealed a G → A point mutation at nt 1062, which should create a stop codon, leading to a truncated protein that is 37% of the wild-type length. The lpm25 mutant was phenotypically indistinguishable from a deletion strain (data not shown).
Low levels of yeast telomerase RNA in mtr10 cells.
We examined telomere length in mtr10Δ cells that had been successively subcultured (Fig. 2A). We observed a progressive reduction in telomere length as the number of cell divisions increased, which was similar to, but perhaps slower than, the shortening observed in a parallel experiment for tlc1. In contrast, a yku80Δ strain displayed a sudden telomere shortening that then remained stable throughout the course of the experiment (data not shown). Thus, like the telomeres of est1, est2, est3, and tlc1 mutants (for example, see reference 18), the telomeres of mtr10 mutants shorten only gradually with an increasing number of cell divisions (Fig. 2A).
However, unlike est1, est2, est3, and tlc1, the mtr10 mutants did not display senescence, nor did telomeres shorten indefinitely. Instead, mtr10 cells often displayed amplification of Y′ subtelomeric regions (Fig. 1B). The telomere maintenance defect in mtr10 mutants is not complete (see below) and, consequently, telomere shortening may be slower than in the case of est1, est2, etc. This may increase the probability of Y′ amplification, as opposed to senescence.
Since mtr10 mutants behaved in many respects like est1, est2, est3, tlc1, and cdc13, we asked whether overexpression of the latter genes from high-copy-number plasmids could suppress the telomere maintenance defect of mtr10 cells. Overexpression of TLC1 restored wild-type telomere length in mtr10Δ cells, whereas no significant changes were observed with the other genes assayed (Fig. 2B). (Some slight lengthening took place with EST2, but this also occurred in the MTR10 control.) On the other hand, neither TLC1 nor any of the other genes had any effect on the slow growth or Ts lethality of mtr10 mutants, suggesting that telomere shortening is not an indirect effect of the other phenotypes of mtr10 strains.
The suppression of the mtr10 telomere phenotype by TLC1 prompted us to examine the level of Tlc1 RNA in the mtr10 mutant. Northern blotting showed that levels of Tlc1 were very low, whereas other RNAs (ACT1 mRNA, snR30, scR1, and U14) were not significantly affected (Fig. 2C and data not shown). Importantly, a high-copy-number TLC1 plasmid greatly restored (although not completely) the levels of mature Tlc1 to wild-type in an mtr10Δ background (Fig. 2C), providing a sufficient explanation for the suppression of telomere shortening. We conclude that levels of yeast telomerase RNA are greatly and specifically reduced in mtr10 mutants and that the reduced levels of Tlc1 largely account for the short telomere phenotype.
Tlc1 occurs in two forms, a precursor poly(A)+ species, which is the direct product of RNA polymerase II transcription, and a shorter, mature form that lacks the poly(A) tail (mTlc1) (4). In the wild-type background, the endogenous levels of the precursor, poly(A)+ Tlc1 account for only ≈5 to 10% of total Tlc1 (4) and are hardly affected by Mtr10p dysfunction. In fact, in some instances, this form was more readily detectable in the mutant than in the wild-type strain (Fig. 2C). By contrast, the level of mTlc1 was reduced about 14-fold (Fig. 3). Thus, the mtr10 mutant is particularly defective in generating the mature, processed form of Tlc1.
FIG. 3.
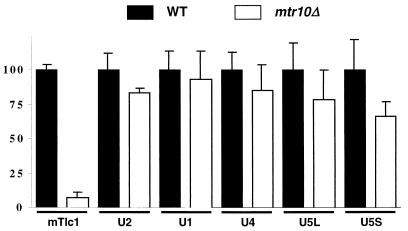
Normal levels of UsnRNAs in mtr10Δ mutants. Equal amounts of total RNA from five wild-type and five mtr10Δ isogenic strains were analyzed by Northern blotting, using probes specific for U1, U2, U4, U5, scR1, and Tlc1. Quantitation was performed using a PhosphorImager and referred to the levels of scR1. Wild-type levels were assigned a value of 100. The means of the five measures and the standard deviations (error bars) are shown.
Normal levels of UsnRNAs in mtr10Δ cells.
Yeast telomerase contains the Sm proteins common to all other snRNPs (45, 55). Association of the Sm proteins with Tlc1 and the UsnRNAs is required for their accumulation (39, 41, 45). In particular, disruption of the interaction between Tlc1 and the Sm proteins leads to phenotypes similar to those described here; that is to say, a reduction in Tlc1 levels and a concomitant decrease in telomere length (45). Thus, Mtr10p might influence this interaction, for instance by importing the Sm proteins to the nucleus, where the snRNPs ultimately function. If a central step in the biogenesis of the snRNPs involving Sm proteins is somehow affected, the levels of other snRNAs should also be low in mtr10 cells.
We therefore compared the levels of UsnRNAs to those of Tlc1 in mtr10Δ and wild-type strains. Little change was observed for any UsnRNA in mtr10 cells (Fig. 3), which argues against a global disturbance in Sm protein function. However, since the processing, maturation, and behavior of UsnRNAs may be quite different from that of Tlc1, we cannot rule out the possibility that some aspects of Sm protein functionality, upon which Tlc1 accumulation might depend to a greater extent, could be affected in an mtr10 mutant.
Role of known cargoes of Mtr10.
Mtr10 is a karyopherin-β, and it helps import proteins into the nucleus. Its major known cargo is the RNA-binding protein Npl3, and other suspected or possible cargoes include the RNA-binding proteins Hrb1 and Gbp2 (38, 44) (Gbp2 only because of its high sequence identity to Hrb1). Furthermore, Gbp2 is a putative telomere-binding protein (16, 19). One of these RNA-binding proteins could mediate an interaction between Mtr10p and Tlc1. However, high-copy overexpression of neither NPL3 nor HRB1 had any effect on the growth rate or telomere length of mtr10 mutants (data not shown).
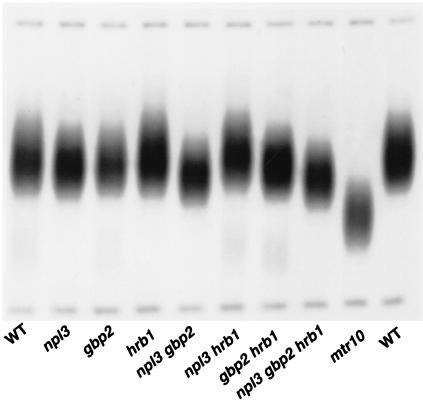
The possible involvement of Npl3, Hrb1, and Gbp2 in telomere metabolism was further addressed by examination of telomere length in single, double, and triple npl3Δ hrb1Δ gbp2Δ mutants (Fig. 4). NPL3 is essential in some genetic backgrounds, but it confers only a growth defect in S288C strains. None of the single mutants exhibited a perceptible alteration in telomere length. A double mutant npl3Δ gbp2Δ displayed a slight telomere shortening that was also observed in the triple mutant hrb1Δ gbp2Δ npl3Δ but not in the other double mutants (Fig. 4). Thus, the Npl3 and Gbp2 proteins might have a role in telomere metabolism. However, this telomere defect is much less severe than in mtr10 mutants.
FIG. 4.
Telomere length in npl3, hrb1, and gbp2 mutants. Telomere length in single, double, and triple mutants as well as wild-type (WT) strains derived from a heterozygous diploid npl3 hrb1 gbp2/NPL3 HRB1 GBP2 was analyzed by Southern blotting. Cells were streaked out for four successive days before DNA was isolated. For comparison, we also included an mtr10Δ mutant in the same genetic background. Only the Y′-type telomeres and size markers are shown.
Defective telomerase biogenesis in mtr10 cells.
The reduced levels of mTlc1 in mtr10 cells could result from defects in transcription of the TLC1 gene or defects in posttranscriptional processing to the mature form, or it could result from abnormally high instability of Tlc1. Two observations suggest that transcription is not defective. Firstly, we were often able to see the poly(A)+ form of Tlc1 in mtr10 cells, sometimes even at higher levels than in a wild-type background (Fig. 2C). Secondly, overexpression of TLC1 gave similarly high levels of the primary poly(A)+ transcript in both wild-type and mtr10 strains, but it gave much higher levels of mature Tlc1 in the wild-type strain than in the mtr10 strain (Fig. 2C and 5B).
To examine the stability of mTlc1, we used a plasmid expressing TLC1 from the regulatable GAL promoter (pGAL-TLC1). In addition, we used mtr10-7, a Ts allele of MTR10 (44), to allow regulatable function of Mtr10. Even at 18°C, mtr10Δ cells carrying the mtr10-7 allele showed lower levels of mTlc1 than MTR10 cells, consistent with only a partial recovery of telomere length in mtr10-7 cells (data not shown). However, significant quantities of mTlc1 accumulated in the mtr10-7 background.
MTR10 tlc1Δ and mtr10Δ tlc1Δ cells carrying pGAL-TLC1 and pRS314-mtr10 were grown at 18°C (permissive temperature) (44) in galactose-containing medium, where expression of TLC1 was high. Both cultures accumulated mTlc1. The cultures were then shifted to 37°C for 2 h to inactivate Mtr10-7 (44), and then glucose was added to repress TLC1 transcription. Levels of mTlc1 were measured over time by Northern analysis (Fig. 5A). After normalization to the initial mTlc1 levels (Fig. 5A, 30 min) and to a loading control (Fig. 5A, scR1 RNA), there was no significant difference between the mtr10-7 and MTR10 strains in the rate of decay of mTlc1; after 90 min, only ≈20% of mTlc1 remained in the MTR10 strain, compared to ≈27% in mtr10Δ mutants (Fig. 5A, 120 min versus 30 min). This result indicates that the mtr10 mutation does not affect mTlc1 stability.
A similar approach was used to study mTlc1 biogenesis. MTR10 tlc1Δ and mtr10Δ tlc1Δ cells carrying pGAL-TLC1 were grown overnight in raffinose medium (noninducing but nonrepressing), then supplemented with galactose to induce TLC1 expression, and sampled at different times. As before, RNA levels were examined by Northern analysis. In the MTR10 tlc1Δ strain both poly(A)+ and poly(A)− forms of Tlc1 were readily apparent, although the appearance of the poly(A)− form was delayed with respect to the poly(A)+ species (Fig. 5B), consistent with a product-precursor relationship (4). On the other hand, the mtr10Δ tlc1Δ strain lacked mTlc1 and had reduced levels of the poly(A)+ form compared to those in MTR10 tlc1Δ cells. Importantly, failure to produce mTlc1 in the mtr10Δ strains was not due to insufficient accumulation of the precursor poly(A)+ Tlc1, because similar amounts of this RNA species did generate mTlc1 in the MTR10 background. Taken together, these results suggest that the absence of Mtr10p disrupts an intermediate step in the biogenesis of Tlc1 and somehow prevents the processing of poly(A)+ Tlc1 into mTlc1.
Tlc1 RNA is mislocalized in mtr10 cells.
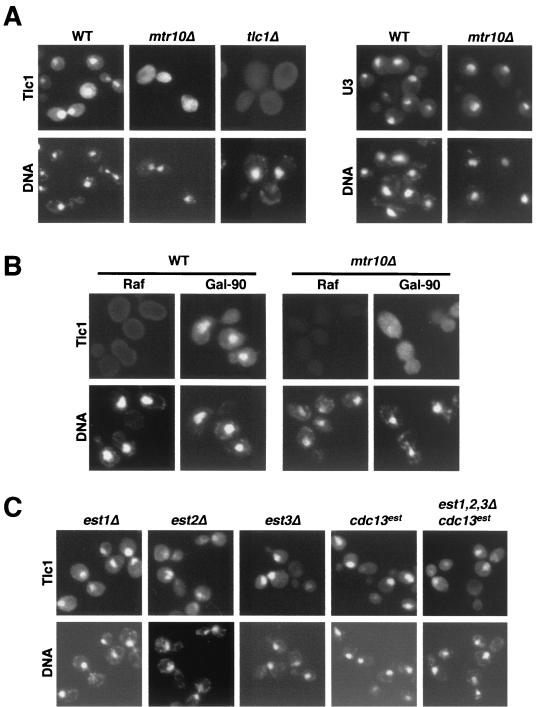
Because Mtr10p functions in nucleocytoplasmic transport, we used RNA FISH to ask whether Tlc1 localization was abnormal in mtr10 mutants. Endogenous levels of Tlc1 were undetectable, so Tlc1 was overexpressed, both from its own promoter (on a high-copy-number plasmid) and from the GAL promoter, and both overexpression methods gave similar results. In a wild-type strain, Tlc1 was mostly nuclear, although some cytoplasmic signal was also apparent (Fig. 6A and B). In mtr10Δ mutants, fewer cells had a detectable signal, as expected from the lower levels of Tlc1, but those cells that did have signal exhibited a dramatic change in Tlc1 localization, such that Tlc1 now appeared dispersed throughout the entire cell (Fig. 6A and B) and was not concentrated in the nucleus. This result suggests that Tlc1 may shuttle through the cytoplasm and that Mtr10p may be involved in its reimport to the nucleus, although alternative explanations also exist (see Discussion).
FIG. 6.
Tlc1 is mislocalized in mtr10Δ cells. (A) Localization of Tlc1 and U3 in wild-type and mtr10Δ strains. Overexpressed Tlc1 (pRS426-TLC1) and endogenous U3 were visualized with antisense RNA probes labeled with Alexa Fluor 488. tlc1Δ cells served as control for Tlc1-specific detection. U3 localization was nuclear in both backgrounds, whereas Tlc1 dramatically changed its localization in the absence of Mtr10p. Here and below, DAPI-stained DNA is also shown. (B) Localization of galactose-induced Tlc1. Cells were grown in raffinose-containing medium, then split in two, and half were supplemented with 2% galactose for 90 min. Tlc1 was visualized with the same probe as above. (C) Localization of Tlc1 in est mutants. est1Δ, est2Δ, and the quadruple mutant were transformed with pGAL-TLC1 and grown overnight in galactose-containing medium. est3Δ and cdc13est carried pRS426-TLC1 and were grown in glucose-containing medium. Tlc1 was visualized as described above. Cells carrying pGAL-TLC1 showed no signal when grown in glucose-containing medium (data not shown).
The dispersed localization was not due to nuclear envelope leakiness, because the nucleolar RNA U3 had normal localization in mtr10 cells (Fig. 6A), further suggesting a rather specific effect of mtr10 on Tlc1. Because pGAL-TLC1 does not give rise to detectable amounts of mTlc1 in mtr10 cells (Fig. 2C and 5B), we attribute the dispersed signal from pGAL-TLC1 mostly to the poly(A)+ form.
In striking contrast, strains deleted for various protein components of telomerase showed normal localization of the RNA subunit (Fig. 6C). Even a quadruple est1Δ est2Δ est3Δ cdc13est mutant exhibited a clear nuclear signal for Tlc1. Furthermore, Tlc1 levels remained fairly constant in these mutants (14, 57; this work [data not shown]).
DISCUSSION
We have shown that levels of Tlc1 drop drastically in the absence of Mtr10p. This is the most likely cause of the telomere shortening observed in mtr10 cells, because overexpression of TLC1 in the mutant restored both telomere length and levels of mTlc1 without suppressing other aspects of the mtr10 phenotype. Because levels of the UsnRNAs remained unchanged in the mtr10Δ strains, it seems that Mtr10p participates in a step in Tlc1 biogenesis not shared by these UsnRNAs.
The mtr10 mutant has particularly low levels of the mature form of Tlc1. Indeed, even when artificially high levels of the precursor form of Tlc1 are provided, mtr10 cells are largely defective in the accumulation of mature Tlc1. This is not because mTlc1 is particularly unstable in mtr10 cells, and so we believe that Mtr10p is somehow important for the processing of poly(A)+ Tlc1 into mTlc1.
Since Mtr10 is a karyopherin, it might mediate the transport of Tlc1. To address this possibility, we studied the subcellular localization of Tlc1. In a wild-type strain, where most Tlc1 is in the mature form, Tlc1 signal was mainly nuclear. In contrast, in an mtr10 GAL-TLC1 strain, where almost all of the Tlc1 is in the poly(A)+ form, most of the Tlc1 was in the cytoplasm, with no detectable nuclear concentration. Thus, the formation of mTlc1 and its appearance in the nucleus are correlated with the presence of functional Mtr10p.
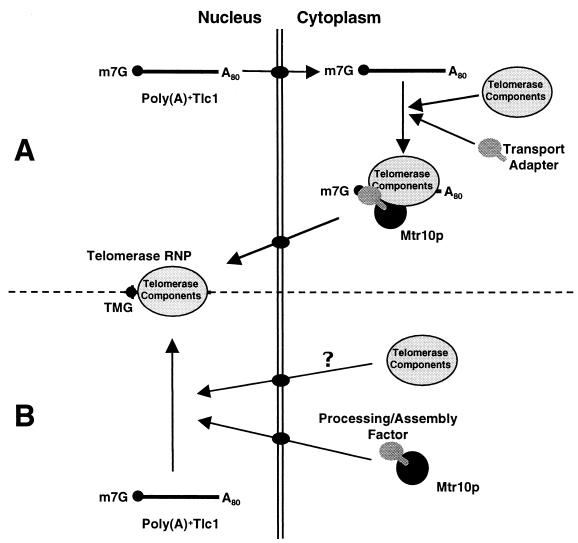
These facts suggest two plausible models for the genesis of mature Tlc1. In the shuttling model (Fig. 7A), the fact that Tlc1 can be seen in the cytoplasm in both wild-type and mtr10 cells is taken to suggest that, after transcription in the nucleus, the poly(A)+ form of Tlc1 is exported to the cytoplasm. Although we know nothing about this putative export, the precursor form of Tlc1 most likely acquires a 5′ m7G cap cotranscriptionally and is polyadenylated (4), so it may follow a general mRNA export pathway. In the cytoplasm, Tlc1 could become associated with some components of telomerase. This Tlc1-protein complex would then be imported into the nucleus via Mtr10p, and the import step would be accompanied by processing of poly(A)+ Tlc1 to mTlc1. For instance, import of the complex might allow binding of the Sm proteins to Tlc1 in the nucleus, triggering the hypermethylation of the cap and the trimming of the 3′ end of poly(A)+ Tlc1 (45, 55). It is not known how the telomerase components Est1p, Est2p, and Est3p are imported into the nucleus, and one appealing possibility is that they are carried in with Tlc1. Note that the converse cannot be true, because Tlc1 localization was not affected by the absence of these proteins. Studies on the localization of Est1, -2, and -3 in tlc1Δ cells should help address this issue.
FIG. 7.
Models for the biogenesis of yeast telomerase. (A) Shuttling model. Tlc1 would travel to the cytoplasm where it would probably pick up some proteins. Its nuclear reimport would be mediated by Mtr10p. Once in the nucleus the processing of Tlc1 would continue with the assembly of the Sm proteins, which would trigger the cap hypermethylation and 3′ end trimming. Note that the Sm proteins would follow a different transport pathway (2). (B) Processing enzyme model. Tlc1 may or may not travel to the cytoplasm, but its reimport would not depend on Mtr10p. Instead, Mtr10p is transporting to the nucleus an enzyme performing an essential function for proper telomerase biogenesis. A80 denotes the poly(A)+ tail of Tlc1 (≈80 nt long) (4).
Whether or not telomerase proteins accompany Tlc1 into the nucleus, it is likely that Mtr10p would require an adapter to interact with Tlc1, since Mtr10 amino acid sequence inspection did not reveal an RNA recognition motif. Interestingly enough, the two identified Mtr10p cargoes, Npl3p and Hrb1p, are RNA-binding proteins (38, 44). The double mutant npl3Δ gbp2Δ had slightly but consistently short telomeres. Given that other yeast proteins also share extensive sequence similarity with Npl3p and Gpb2p, it is plausible that a broader functional redundancy exists.
The full assembly of telomerase may be a difficult process, and it is possible that shuttling from one compartment to another, allowing stepwise, partial assembly, may be a common theme for orchestrating the assembly of many large complexes. Interestingly, in human cells telomerase is able to shuttle between nucleus and cytoplasm in a way that depends on the exportin CRM1p and members of the 14-3-3 protein family (42). However, the in vivo significance of this nucleocytoplasmic shuttling is yet to be determined.
In the processing enzyme model (Fig. 7B), Mtr10p would be involved in the nuclear import of a processing or assembly factor essential for Tlc1 maturation. In the absence of this factor, the overproduced and unprocessed Tlc1 would be aberrantly exported to the cytoplasm by the general mRNA export machinery, where it would be degraded. The putative processing factor imported by Mtr10p might have a function similar to that of Brr1p; brr1Δ cells have low levels of UsnRNAs because they are deficient in an early step in UsnRNA biogenesis (32). These authors found that a truncated U2 snRNA presents a 3′-extended precursor that is rapidly processed into the mature form. However, in the absence of Brr1p, this process is greatly delayed. The factor is not Brr1p in this case, however, because Tlc1 levels were normal in a brr1Δ background (data not shown).
In higher eukaryotes, UsnRNAs transit the cytoplasm, where they bind the Sm proteins. This binding triggers, among other processes, the trimming of the 3′ end of these RNAs (55). It has been proposed that Tlc1, a UsnRNA-like RNA, might follow a similar step in its biogenesis (45), although it is unknown whether UsnRNAs in yeast transit the cytoplasm or not. Our data are very suggestive that telomerase biogenesis may involve a cytoplasmic phase for Tlc1. However, our data are most consistent with a model in which the trimming of the poly(A) tail of Tlc1 occurs in the nucleus, which supports the hypothesis that in yeast the Sm proteins follow an RNA-independent nuclear import pathway (2).
The processing, assembly, and transport of the yeast telomerase RNP is complex, and there are many possibilities for defects causing problems in telomere maintenance. The same is undoubtedly true in mammals. For example, a human genetic disease, dyskeratosis congenita, appears to be the result of a defect in telomerase. The telomerase defect can occur as a result of mutations in the gene for the human telomerase RNA, hTR (52), or as a result of mutations in the dyskerin gene (28), which encodes a protein involved in the processing of hTR and of other RNAs. The molecular processes affected by Mtr10 in yeast, and dyskerin in humans, are quite different, and each is potentially capable of affecting a wide range of cellular functions. Nevertheless, it is striking that these defects give strong telomere-related phenotypes. This speaks to the importance of proper assembly and processing of telomerase in widely diverged organisms.
Acknowledgments
We thank the following labs for sending us materials: Ares, Blobel, Cameron, Cech, Charbonneau, Cole, Elledge, Fabre, Gottschling, Guthrie, Haber, Hartwell, Herskowitz, Hieter, Hurt, Iggo, Jackson, Jacobson, Johnson, Kaufman, Kleckner, Lin, Lundblad, Nishizawa, Ogawa, Petes, Rine, Rose, Rothstein, Runge, Seraphin, Shore, Silver, Strathern, Wigler, and Zakian. F.F. thanks former and current members of the Futcher lab for helpful discussions and technical help.
This work was funded by grant DAMD17-97-1-7315 from the Army Breast Cancer program and in part by grants from the National Institutes of Health. F.F. was supported by a fellowship from the Ministerio de Educación y Cultura, Spain.
REFERENCES
- 1.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 2.Bordonné, R. 2000. Functional characterization of nuclear localization signals in yeast Sm proteins. Mol. Cell. Biol. 20:7943-7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, T. 1994. Preparation and analysis of DNA, p. 2.9.7-2.9.8. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. More, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 4.Chapon, C., T. R. Cech, and A. J. Zaug. 1997. Polyadenylation of telomerase RNA in budding yeast. RNA 3:1337-1351. [PMC free article] [PubMed] [Google Scholar]
- 5.Collart, M. A., and S. Oliviero. 1994. Saccharomyces cerevisiae, p. 13.12.1-13.12.2. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. More, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 6.Conti, E., and E. Izaurralde. 2001. Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 13:310-319. [DOI] [PubMed] [Google Scholar]
- 7.Counter, C. M., M. Meyerson, E. N. Eaton, and R. A. Weinberg. 1997. The catalytic subunit of yeast telomerase. Proc. Natl. Acad. Sci. USA 94:9202-9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elela, S. A., and M. Ares, Jr. 1998. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 17:3738-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 10.Görlich, D., M. Dabrowski, F. R. Bischoff, U. Kutay, P. Bork, E. Hartmann, S. Prehn, and E. Izaurralde. 1997. A novel class of RanGTP binding proteins. J. Cell Biol. 138:65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 12.Greider, C. W. 1996. Telomere length regulation. Annu. Rev. Biochem. 65:337-365. [DOI] [PubMed] [Google Scholar]
- 13.Huber, J., U. Cronshagen, M. Kadokura, C. Marshallsay, T. Wada, M. Sekine, and R. Lührmann. 1998. Snurportin-1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 17:4114-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, T. R., S. K. Evans, R. G. Weilbaecher, and V. Lundblad. 2000. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 10:809-812. [DOI] [PubMed] [Google Scholar]
- 15.Kadowaki, T., S. Chen, M. Hitomi, E. Jacobs, C. Kumagai, S. Liang, R. Schneiter, D. Singleton, J. Wisniewska, and A. M. Tartakoff. 1994. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol. 126:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konkel, L. M. C., S. Enomoto, E. M. Chamberlain, P. McCune-Zierath, S. J. P. Iyadurai, and J. Berman. 1995. A class of single-stranded telomeric DNA-binding proteins required for Rap1p localization in yeast nuclei. Proc. Natl. Acad. Sci. USA 92:5558-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, M. S., M. Henry, and P. A. Silver. 1996. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 10:1233-1246. [DOI] [PubMed] [Google Scholar]
- 18.Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian, and V. Lundblad. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144:1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, J., and V. A. Zakian. 1994. Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1-3)n single strand telomeric DNA in vitro. Nucleic Acids Res. 22:4906-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, J., and V. A. Zakian. 1995. An in vitro assay for Saccharomyces telomerase requires EST1. Cell 81:1127-1135. [DOI] [PubMed] [Google Scholar]
- 21.Lingner, J., J. P. Cooper, and T. R. Cech. 1995. Telomerase and DNA end replication: no longer a lagging strand problem? Science 269:1533-1534. [DOI] [PubMed] [Google Scholar]
- 22.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 23.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 24.Lundblad, V. 1994. Saccharomyces cerevisiae, p. 13.10.5-13.10.6. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. More, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 25.Lundblad, V., D. A. Treco, F. Winston, and D. M. Becker. 1994. Saccharomyces cerevisiae, p. 13.1.1-13.3.4 and 13.7.1-13.7.10. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. More, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 26.Lustig, A. J. 1992. Hoogsteen G-G base pairing is dispensable for telomere healing in yeast. Nucleic Acids Res. 20:3021-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Mitchell, J. R., E. Wood, and K. Collins. 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402:551-555. [DOI] [PubMed] [Google Scholar]
- 29.Moy, T. I., and P. A. Silver. 1999. Nuclear export of the small ribosomal subunit requires the Ran-GTPase cycle and certain nucleoporins. Genes Dev. 13:2118-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakielny, S., U. Fischer, W. M. Michael, and G. Dreyfuss. 1997. RNA transport. Annu. Rev. Neurosci. 20:269-301. [DOI] [PubMed] [Google Scholar]
- 31.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 32.Noble, S. M., and C. Guthrie. 1996. Transcriptional pulse-chase analysis reveals a role for a novel snRNP-associated protein in the manufacture of spliceosomal snRNPs. EMBO J. 15:4368-4379. [PMC free article] [PubMed] [Google Scholar]
- 33.Nugent, C. I., and V. Lundblad. 1998. The telomerase reverse transcriptase: components and regulation. Genes Dev. 12:1073-1085. [DOI] [PubMed] [Google Scholar]
- 34.Ohno, M., A. Segref, A. Bachi, M. Wilm, and I. W. Mattaj. 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101:187-198. [DOI] [PubMed] [Google Scholar]
- 35.Olovnikov, A. M. 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41:181-190. [DOI] [PubMed] [Google Scholar]
- 36.O'Reilly, M., S. A. Teichmann, and D. Rhodes. 1999. Telomerases. Curr. Opin. Struct. Biol. 9:56-65. [DOI] [PubMed] [Google Scholar]
- 37.Palacios, I., M. Hetzer, S. A. Adam, and I. W. Mattaj. 1997. Nuclear import of U snRNPs requires importin beta. EMBO J. 16:6783-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pemberton, L. F., J. S. Rosenblum, and G. Blobel. 1997. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J. Cell Biol. 139:1645-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy, J., B. Zheng, B. C. Rymond, and J. L. Woolford, Jr. 1995. Structurally related but functionally distinct yeast Sm D core small nuclear ribonucleoprotein particle proteins. Mol. Cell. Biol. 15:445-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell, I. D., and D. Tollervey. 1992. NOP3 is an essential yeast protein which is required for pre-rRNA processing. J. Cell Biol. 119:1077-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rymond, B. C. 1993. Convergent transcripts of the yeast PRP38-SMD1 locus encode two essential splicing factors, including the D1 core polypeptide of small nuclear ribonucleoprotein particles. Proc. Natl. Acad. Sci. USA 90:848-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seimiya, H., H. Sawada, Y. Muramatsu, M. Shimizu, K. Ohko, K. Yamane, and T. Tsuruo. 2000. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 19:2652-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seipelt, R. L., B. Zheng, A. Asuru, and B. C. Rymond. 1999. U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 27:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senger, B., G. Simos, F. R. Bischoff, A. Podtelejnikov, M. Mann, and E. Hurt. 1998. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J. 17:2196-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seto, A. G., A. J. Zaug, S. G. Sobel, S. L. Wolin, and T. R. Cech. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401:177-180. [DOI] [PubMed] [Google Scholar]
- 46.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 48.Stage-Zimmermann, T., U. Schmidt, and P. A. Silver. 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11:3777-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steiner, B. R., K. Hidaka, and B. Futcher. 1996. Association of the Est1 protein with telomerase activity in yeast. Proc. Natl. Acad. Sci. USA 93:2817-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Virta-Pearlman, V., D. K. Morris, and V. Lundblad. 1996. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 10:3094-3104. [DOI] [PubMed] [Google Scholar]
- 52.Vulliamy, T., A. Marrone, F. Goldman, A. Dearlove, M. Bessler, P. J. Mason, and I. Dokal. 2001. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413:432-435. [DOI] [PubMed] [Google Scholar]
- 53.Watson, J. D. 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239:197-201. [DOI] [PubMed] [Google Scholar]
- 54.Will, C. L., and R. Lührmann. 1997. snRNP structure and function, p. 130-173. In A. R. Krainer (ed.), Eukaryotic mRNA processing. IRL Press, Oxford, United Kingdom.
- 55.Will, C. L., and R. Lührmann. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290-301. [DOI] [PubMed] [Google Scholar]
- 56.Winston, F. 1994. Saccharomyces cerevisiae, p. 13.3.1-13.3.2. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. More, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 57.Zhou, J., K. Hidaka, and B. Futcher. 2000. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol. Cell. Biol. 20:1947-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]