FIG.1.
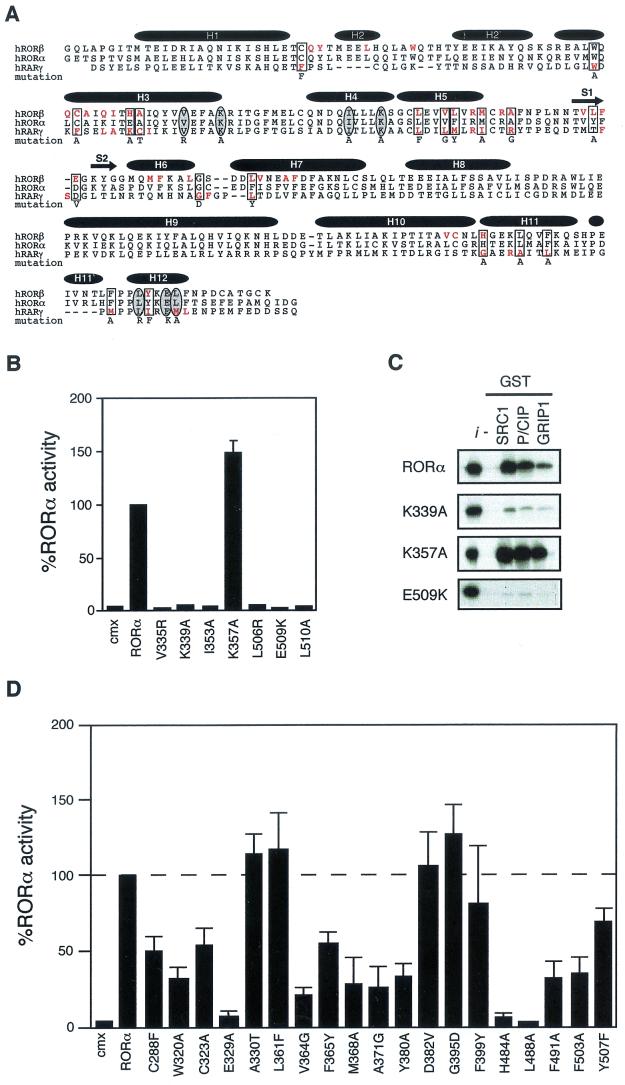
RORα shares common structural and functional determinants with classic nuclear receptors. (A) Primary sequence of RORβ, RORα, and RARγ ligand binding domains. Amino acids involved in the LBP identified by crystallographic analysis are highlighted in red. Amino acids essential for AF-2 activity and known to participate in ligand binding targeted for site-directed mutagenesis are circled and boxed, respectively. The respective amino acid change is indicated below the sequence. The secondary structure is represented by black bars for the α-helices and arrows for the β-sheets. (B) RORα hydrophobic cleft mutants (V335R, K339A, and I353A) and AF-2 helix mutants (L506R, E509K, and L510A) are transcriptionally inactive in transfected Cos-1 cells, with the exception of the cleft mutant K357A. Normalized values are calculated in terms of percent RORα activity with respect to wild type. These results are the average of three independent experiments. (C) Binding of RORα and hydrophobic cleft (K339A, K357A) and AF-2 helix (E509K) mutants to SRC proteins. GST-SRC1aRID, GST-p/CIPRID, and GST-GRIP1RID fusion proteins were coupled to Sepharose beads incubated with 35S-labeled RORα, RORαK339A, RORαK357A, and RORαE509K. The input lane (i) represents 10% of total lysate included in the binding reaction. (D) Cos-1 cells were cotransfected with RORα LBP mutants and ROREα23-TkLuc reporter. Normalized luciferase values are expressed as percent activity with respect to wild-type RORα. These results are the average of three independent experiments.