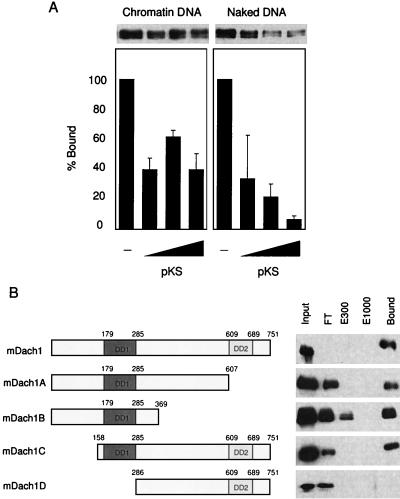
FIG. 5.
Binding of Dach1 to naked and chromatin DNA. (A) One hundred nanograms each of chromatin DNA-conjugated Dynabeads (left panel) and naked DNA-conjugated beads (right panel) was incubated with the nuclear extract of NIH 3T3 cells transfected with pfDach1 and washed, and then an increasing amount of pBluescript KS (1, 5, and 25 μg) was added and incubated. After washing, FLAG-Dach1 protein was analyzed by Western blotting (upper panel). The relative amount of FLAG-Dach1 protein was quantitated by densitometry, and those without pBluescript KS (columns 1 and 5) were set at 100. Three exposures were quantitated, and the average and standard deviation are shown. (B) pfDach1 and its deletion constructs (the region in each construct is shown in the left panel) were transfected into NIH 3T3 cells, and nuclear extracts of these cells were incubated with 50 μl of natural DNA cellulose. After centrifugation, supernatant was collected as flowthrough (FT). DNA cellulose beads were eluted in a buffer containing 300 mM NaCl (E300). After washing in the buffer containing 300 mM NaCl, beads were eluted in a buffer containing 1,000 mM NaCl (E1000). Each supernatant and proteins bound to the resin after a washing in 1,000 mM NaCl (Bound) were analyzed by Western blotting using anti-FLAG antibody.