Figure 2.
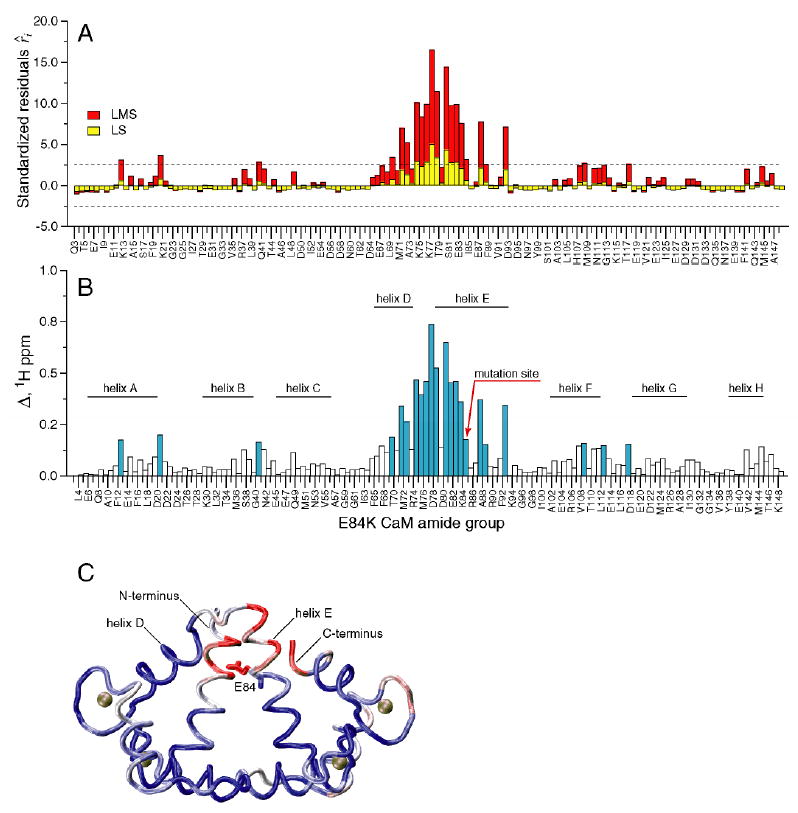
Chemical shift-based identification of structural perturbations in the CaM-smMLCKp complex introduced by the E84K mutation in calmodulin. (A) Standardized residuals calculated using one-dimensional LMS (red) and LS (yellow) analyses of 15N-1H cross-peak displacements Δ in the E84K-smMLCKp complex. Dotted lines represent the ± 2.5 interval of the standardized residuals. The amide groups that have absolute values of the residuals greater than 2.5 are considered to be chemical shift outliers. (B) 15N-1H cross-peak displacement Δ in the E84K-smMLCKp complex. The amide groups that are identified as outliers by the LMS analysis are colored blue. Positions of the CaM helices are indicated by the solid bars. 22 outliers are identified by LMS, compared to only 7 outliers identified by conventional LS. (C) Squared displacement of backbone atoms between the wild-type CaM-smMLCKp (PDB code 1cdl) and E84K-RS20 (PDB code 1vrk) structures mapped onto the structure of CaM-smMLCKp. The structure of the peptide is not shown. A blue-white-red color gradient is used to represent the range of squared atom displacements from 0 to 3 Å2. The average R.M.S.D. for backbone atoms of residues 5-146 of calmodulin is 0.94 Å. The results of superposition of the CaM-smMLCKp and E84K-RS20 structures are in general consistent with amide chemical shift data, where helix E and the loop between helices E and D can be identified as the most perturbed protein regions.
