Figure 4.
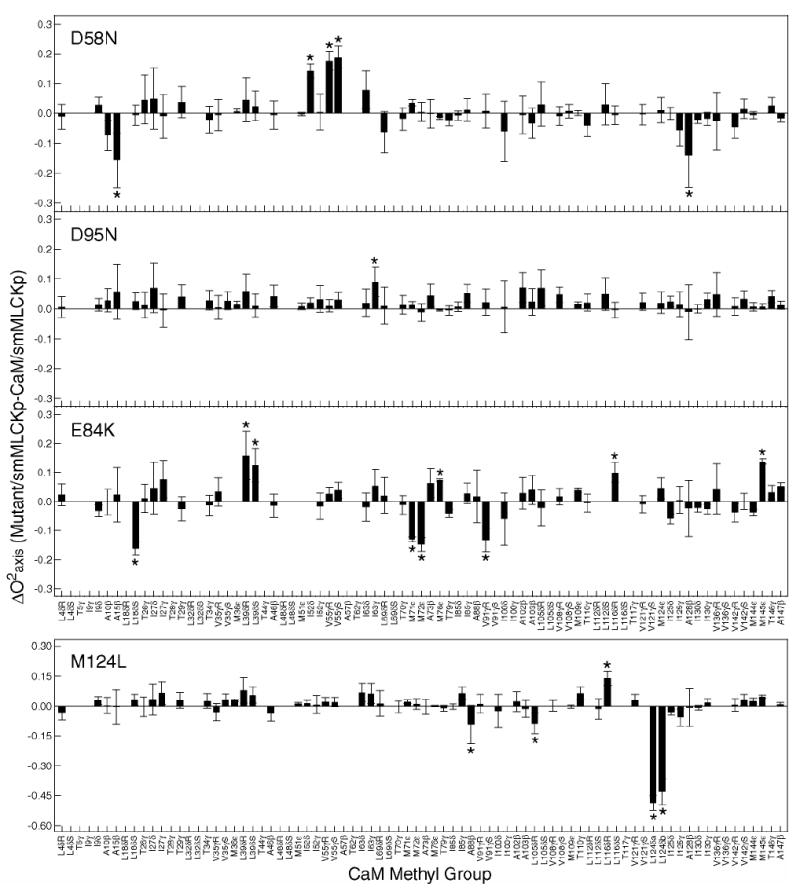
Identification of dynamical perturbations in the CaM-smMLCKp complex introduced by various mutations in calmodulin. Difference between methyl group symmetry axis order parameters of the mutant and wild-type smMLCKp complexes. The errors bars are calculated as a sum of individual absolute errors of the corresponding mutant and wild-type order parameters. Outliers identified by LMS analysis are labeled with asterisks. Three patterns of perturbation in the side chain methyl dynamics are observed: almost no perturbation in D95N-, mostly local in M124L-, and long-range in D58N- and E84K-smMLCKp complexes. Note that the vertical and horizontal scales in the M124L graph are different from those in the other three graphs.
