Figure 7.
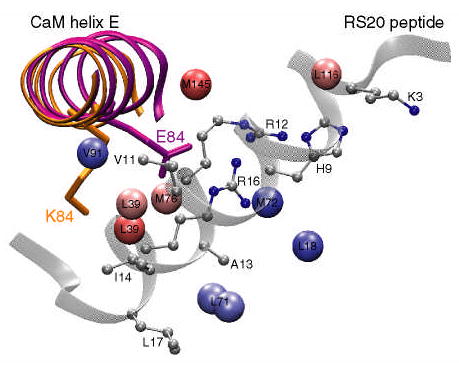
A cartoon of the E84K-RS20 structure (PDB code 1vrk) showing the relative position of calmodulin helix E and the RS20 peptide. The structures of helix E in the wild-type and E84K complexes are superimposed and shown in purple and orange, respectively. The mutated side chain is shown in stick representation and is labeled. In the E84K-RS20 complex, the side chain of K84 rotates away from the peptide. The values of standardized residuals are mapped onto methyl carbons using the same color gradient as in Figure 6. Since in VU-1 CaM position 71 is occupied by leucine, the standardized residual of M71ɛ is mapped onto the leucine δ methyl groups. The peptide side chains that are within 5 Å of the perturbed methyls are shown in ball-and-stick representation.
