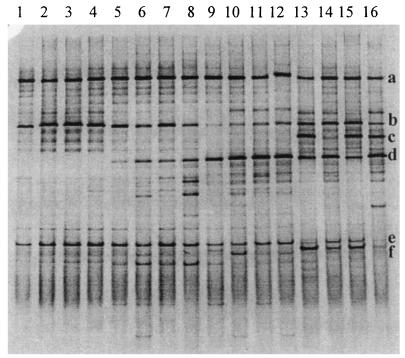
FIG. 2.
PCR-DGGE profiles (30 to 55% denaturing gradient gel) generated from representative ileal content pools, using primer pair HDA1-GC plus HDA2. Lanes: 1 to 4, samples from 7-day-old chickens fed the A− diet (lane 1), the A+ diet (lane 2), the S− diet (lane 3), and the S+ diet (lane 4); 5 to 8, samples from 14-day-old chickens fed the A− diet (lane 5), the A+ diet (lane 6), the S− diet (lane 7), and the S+ diet (lane 8); 9 to 12, samples from 21-day-old chickens fed the A− diet (lane 9), the A+ diet (lane 10), the S− diet (lane 11), and the S+ diet (lane 12); 13 to 16, samples from 35-day-old chickens fed the A− diet (lane 13), the A+ diet (lane 14), the S− diet (lane 15), and the S+ diet (lane 16). The fragments were allotted by sequence analysis to the following genera or species: fragment a, L. johnsonii; fragment b, L. crispatus; fragment c, L. salivarius; fragment d, Streptococcus spp.; fragment e, sequence similarity of 98% to an uncultured bacterium and 97% to L. reuteri; fragment f, C. perfringens.