Abstract
Hexachlorocyclohexane (HCH) has been used extensively against agricultural pests and in public health programs for the control of mosquitoes. Commercial formulations of HCH consist of a mixture of four isomers, α, β, γ, and δ. While all these isomers pose serious environmental problems, β-HCH is more problematic due to its longer persistence in the environment. We have studied the degradation of HCH isomers by Sphingomonas paucimobilis strain B90 and characterized the lin genes encoding enzymes from strain B90 responsible for the degradation of HCH isomers. Two nonidentical copies of the linA gene encoding HCH dehydrochlorinase, which were designated linA1 and linA2, were found in S. paucimobilis B90. The linA1 and linA2 genes could be expressed in Escherichia coli, leading to dehydrochlorination of α-, γ-, and δ-HCH but not of β-HCH, suggesting that S. paucimobilis B90 contains another pathway for the initial steps of β-HCH degradation. The cloning and characterization of the halidohydrolase (linB), dehydrogenase (linC and linX), and reductive dechlorinase (linD) genes from S. paucimobilis B90 revealed that they share ∼96 to 99% identical nucleotides with the corresponding genes of S. paucimobilis UT26. No evidence was found for the presence of a linE-like gene, coding for a ring cleavage dioxygenase, in strain B90. The gene structures around the linA1 and linA2 genes of strain B90, compared to those in strain UT26, are suggestive of a recombination between linA1 and linA2, which formed linA of strain UT26.
Hexachlorocyclohexane (HCH) is a chlorinated cyclic saturated hydrocarbon, also known as benzene hexachloride, or BHC. In the recent past, γ-HCH (lindane) and technical BHC (a mixture of HCH isomers) have been used extensively, particularly for the control of agricultural pests and mosquitoes. Commercially, technical BHC is prepared by chlorination of benzene in the presence of UV light. During its synthesis, primarily four isomers, α, β, γ, and δ, are formed in the ratio 65, 12, 12, and 7%, respectively (36). Among these, only γ-HCH has insecticidal properties, and its isolation requires several concentration and purification steps, thereby increasing its cost. Thus, primarily for economic reasons, technical BHC has been used widely and indiscriminately in several developing countries, including India. Although the use of lindane and BHC has been banned or severely restricted in a number of countries, including India, the ban has not stopped HCH residues from entering the environment. As a result, HCH residues are being detected in various environmental niches, not only in India but all over the world (4, 26, 27, 32). While γ-HCH is persistent in the environment but still degradable, the α and β isomers are the more problematic of the different HCH isomers in view of their very low solubility in water and thus their high potential for bioaccumulation. There is also evidence that the α and γ isomers of HCH are converted into the β isomer in living organisms (7). A survey for HCH residues in various samples conducted over many years showed that most are contaminated with α- and β-HCH (7, 26, 12).
The persistence of HCH isomers in aerobic environments is primarily due to the absence of microbes that can degrade them (8, 10, 11, 19). HCH isomers are known to degrade slowly under anaerobic conditions (10, 14, 28), but there are very few reports of HCH degradation under aerobic conditions. Matsumura et al. (15) first reported aerobic degradation of HCH by a Pseudomonas strain. Degradation of HCH by a Pseudomonas paucimobilis strain (later reclassified as Sphingomonas paucimobilis strain SS86) was reported in upland experimental fields in Japan where γ-HCH had been applied once a year for 12 years (31, 35). γ-HCH-degrading S. paucimobilis was also isolated from French soils (33). Despite the fact that these strains degraded α-, δ-, and γ-HCH, they were unable to degrade β-HCH.
The failure to discover microbes that could degrade β-HCH aerobically (2, 5, 6, 33) suggested that bacteria were unable to degrade β-HCH due to its unique chemical structure and stereochemistry. However, two reports described S. paucimobilis strain B90, isolated in Cuttack, India, from rice rhizosphere soil that had been treated repeatedly with technical BHC (3, 29), which was able to mineralize not only α-, δ-, and γ-HCH but also β-HCH (9, 29).
The genes and enzymes which trigger the degradation of γ-HCH have been characterized mostly from S. paucimobilis UT26 (6, 19, 24), which is a nalidixic acid-resistant mutant of S. paucimobilis strain SS86. The primary enzyme for γ-HCH degradation in strain UT26 is HCH dehydrochlorinase, encoded by the gene linA (6, 22, 25). The remaining genes of the γ-HCH degradative pathway, i.e., linB (21, 23, 25), linC (20, 25), linD (16), and linE (17, 18), encode a halidohydrolase, a dehydrogenase, a reductive dechlorinase, and a dioxygenase, respectively, and have also been cloned and characterized from S. paucimobilis UT26. Parts of the genes for γ-HCH dehydrochlorinase and for the remainder of the HCH degradative pathway were amplified by PCR from DNA of the S. paucimobilis strain isolated from French soil (33), suggesting that HCH degradation is probably mediated by enzymes similar to those of S. paucimobilis UT26.
Since S. paucimobilis strain B90 degrades α-, β-, γ-, and δ-HCH, this organism is promising for developing bioremediation strategies for the decontamination of soils containing HCH residues. As a first step in this direction, we report here the degradation pattern of HCH isomers and the cloning and characterization of several lin genes for the HCH degradative pathway from S. paucimobilis B90.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. S. paucimobilis B90 (29) was obtained from N. Sethunathan, Central Rice Research Institute, Cuttack, India, and S. paucimobilis UT26 (31, 35) was obtained from Y. Nagata (University of Tokyo, Yayoi, Bunkyo-ku, Tokyo, Japan). Plasmids pMIA2, pYNA4R, pBFRU2, and pKM2 contain truncated fragments of the linA, linB, linC, and linD genes of S. paucimobilis UT26, respectively (Table 1), and were also obtained from Y. Nagata. S. paucimobilis B90 and S. paucimobilis UT26 were usually grown at 28°C in mineral salt medium (SM) containing (per liter) 0.5 g of (NH4)2HPO4, 0.2 g of MgSO4 · 7H2O, 0.1 g of K2HPO4, 0.01 g of Ca(NO3)2, and 0.01 g of FeSO4 · 7H2O, supplemented with 1% glucose. Escherichia coli DH5α and JM101 were used as general host strains for plasmid cloning. The E. coli strains were grown routinely at 37°C in Luria broth (LB) or on LB agar (30) supplemented with the appropriate antibiotics when necessary.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Sourceb |
|---|---|---|
| E. coli DH5α | F−endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 ΔU169 deoR | Laboratory stock |
| E. coli BL21 | B F−ompT hsdS (rB− mB−) gal dcm | Amersham Pharmacia |
| E. coli JM101 | supE thi Δ(lac-proAB) F′[tra D36 proAB+lacIqlacZ ΔM15] | Laboratory stock |
| S. paucimobilis B90 | Aerobic, motile, rod shaped; degrades all four isomers of HCH (α, β, γ, and δ) | CRRI, Cuttack, India |
| S. paucimobilis UT26 | Aerobic, motile, rod shaped; degrades α, γ, and δ-HCH but not β-HCH | Y. Nagata |
| pUC 13/19 | 2.7-kb; Ampr; MCS internal to lacZ gene | Laboratory stock |
| pGEX-5X-3 | GST fusion vector with tac promoter, lacIq, factor Xa protease recognition site | Amersham Pharmacia |
| pIMA2 | Recombinant construct carrying truncated linA gene | Y. Nagata |
| pYNA4R | Recombinant construct carrying truncated linB gene | Y. Nagata |
| pBFRU2 | Recombinant construct carrying truncated linC gene | Y. Nagata |
| pKM2 | Recombinant construct carrying truncated linD gene | Y. Nagata |
| pLIN14 | pUC13 carrying 1.4-kb BclI fragment from genomic DNA of S. paucimobilis B90 | This study |
| pLIN25 | pUC13 carrying 2.5-kb BclI fragment from genomic DNA of S. paucimobilis B90 | This study |
| pLINA1 | pUC19 containing linA1 gene | This study |
| pLINA2 | pUC19 containing linA2 gene | This study |
| pLINB | pUC19 containing linB gene | This study |
| pLINC | pUC19 containing linC gene | This study |
| pLIND | pUC19 containing linD gene | This study |
| pLINX | pUC19 containing linX gene | This study |
| pLINEA1 | pGEX-5X-3 containing linA1 gene | This study |
| pLINEA2 | pGEX-5X-3 containing linA2 gene | This study |
MCS, multiple cloning site; GST, glutathione S-transferase.
CRRI, Central Rice Research Institute.
Chemicals and enzymes.
Analytical grade α-, β-, γ-, and δ-HCH were obtained from Ehrenstorfer GmbH (D-86199, Augsburg, Germany). Pentachlorocyclohexene (PCCH) was chemically synthesized from α-, γ-, and δ-HCH according to the method described by Trantirek et al. (34). Enzymes for DNA manipulations were purchased from Bangalore Genei (Bangalore, India), New England Biolabs Inc. (Beverly, Mass.), and Boehringer Mannheim (Mannheim, Germany). The Nonradioactive DNA Labeling and Detection kit was purchased from Boehringer Mannheim, and [α-32P]dATP was obtained from the Bhabha Atomic Research Centre (Mumbai, India).
Analytical techniques.
The degradation of α-, β-, γ-, and δ-HCH in S. paucimobilis B90, S. paucimobilis UT26, and recombinant E. coli containing plasmids with the lin genes was carried out either in SM supplemented with 1% glucose or in LB. Precultures of S. paucimobilis or E. coli grown overnight in SM plus glucose or LB were transferred at 1% (vol/vol) to fresh medium containing 5 μg of each HCH isomer/ml. Each flask containing α-, β-, γ-, or δ-HCH was incubated at 28°C (37°C for E. coli) on a rotary shaker. Appropriate controls containing medium plus the HCH isomer or medium plus the HCH isomer and E. coli containing pUC13-pUC19 or pGEX-5X-3 were kept simultaneously. Aliquots (200 μl) were taken out periodically and extracted twice with 500 μl of hexane. The concentration of HCH isomers was measured by subjecting samples to GC on a chromatograph equipped with an electron capture 63Ni detector (GC 17A; Shimadzu, Kyoto, Japan) on a column containing 3% OV 17 (Chromatopak Analytical Instrumentation Pvt. Ltd, Mumbai, India). The column, injector, and detector temperatures were maintained at 200, 220, and 250°C, respectively, with a flow rate of carrier gas (nitrogen) of 27 ml/min.
DNA manipulation.
Plasmid DNA from E. coli was isolated by the alkaline lysis method (30). The genomic DNA of S. paucimobilis was isolated by standard methods from cells pelleted from 100-ml cultures grown to stationary phase in SM containing 1% glucose (30). Southern hybridizations with fragments of the linA, linB, linC, and linD genes of S. paucimobilis UT26 (Table 1) were carried out by established procedures (30). DNA sequences were determined using standard methodologies for double-stranded plasmid DNA on an automated DNA sequencer (ABI PRISM model 377, version 3; Applied Biosystems) at the Department of Biochemistry, South Campus, University of Delhi, Delhi, India. The sequences were analyzed using the DNASIS package (Pharmacia). The BLAST program (1) was used for homology searches within GenBank, and ClustalW was used for multiple alignments of sequences.
Cloning of lin genes.
The lin genes were cloned from S. paucimobilis B90 as follows. Two BclI fragments of 1.4 and 2.5 kb hybridizing to the linA probe from strain UT26 were eluted from BclI-digested strain B90 genomic DNA and ligated with BamHI-digested pUC13. After ligation and transformation, colonies hybridizing positively to the linA probe were recovered. Genomic DNA of S. paucimobilis B90 was partially digested with Sau3AI and size fractionated on a sucrose density gradient. DNA fragments of 35 to 45 kb were ligated to the BamHI-digested cosmid pWE15 (Stratagene, La Jolla, Calif.). The cosmid library of S. paucimobilis B90 was screened with the linA, linB, linC, and linD gene probes described above. Positive clones were regrown, and the cosmids were isolated. Total genomic DNA from S. paucimobils B90 or cosmid clones containing appropriate inserts were also used as templates for PCR amplification with conserved lin gene primers (Table 2). PCR amplification was performed with a Nugene Thermocycler (Techne, Progene, Cambridge, United Kingdom) according to the specifications of the supplier. The PCR products were cloned in pUC13 and in the E. coli expression vector pGEX-5X-3 (Stratagene) in order to study the possible expression and activities of the lin gene products.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′)a | Added restriction site | Designation | Source or accession no. |
|---|---|---|---|---|
| 1 | GCGGATCCGCATGAGTGATCTAGACAGACTT | BamHI | linA-sense | D90355 |
| 2 | GCCTCGAGTTATGCGCCGGACGGTGCGAAATG | XhoI | linA1-antisense | This study |
| 3 | GCCTCGAGTCACGATTTTTGCAACAGAGC | XhoI | linA2-antisense | This study |
| 4 | GCGGATCCGCATGAGCCTCGGCGCAAAGCCA | BamHI | linB-sense | D14594 |
| 5 | GCCTCGAGTTATGCTGGGCGCAATCGCCGGAC | XhoI | linB-antisense | D14594 |
| 6 | GCGGATCCGCATGTCTGATTTGAGCGGC | EcoRI | linC-sense | D14595 |
| 7 | GCCTCGAGTCAGATCGCGGTAAAGCCGCCGTC | XhoI | linC-antisense | D14595 |
| 8 | GCGAATTCAATGAGCGCTGATACAGAA | EcoRI | linD-sense | D89733 |
| 9 | GCCTCGAGTTAGGCGTTGCTCAGGAGATGGAT | XhoI | linD-antisense | D89733 |
| 10 | GCGGATCCGCATGGCTAACAGACTCGCAGGCA | BamHI | linX-sense | D23722 |
| 11 | GCCTCGAGTCAAACACCCACGGACCAGCCTCC | XhoI | linX-antisense | D23722 |
| 12 | AGGAATTCCATGATGCAACTGCCCGAA | EcoRI | linE-sense | AB021867 |
| 13 | AGCTCGAGCTCAAATGACGATCGGATC | XhoI | linE-antisense | AB021867 |
| 14 | TGGGATCCCCGTGAATATAGATGACCTGG | BamHI | linR-sense | AB021860 |
| 15 | GGGTCGACTCACATCCGCGCGGACAG | SalI | linR-antisense | AB021860 |
| 16 | GCGGATCCGCTCTGTTGCAAAAATCGTGAAGC | BamHI | Tn610-sense | X536535 |
| 17 | GCGGATCCGATGACCATGATTACGCC | BamHI | Tn610-antisense | X536535 |
Restriction sites are underlined.
HCH dehalogenase activity in linA1 and linA2 clones.
E. coli containing pLINEA1 or pLINEA2 was grown in LB plus 50 μg of ampicillin/ml and 5 μg of each HCH isomer/ml. Samples (200 μl) were withdrawn periodically with a micropipette, extracted twice with 500 μl of hexane, and subjected to GC as described above. Cell extracts were prepared from the same E. coli clones grown in 50 ml of LB medium containing ampicillin at an optical density at 600 nm of 1. The cells were harvested by centrifugation at 4,500 × g for 10 min at 4°C. The pellet was resuspended in sonication buffer (0.25 M glucose, 5 mM dithiothreitol, 2 mM Na2EDTA, 150 mM NaCl, 50 mM Tris HCl, 100 mM phenylmethylsulfonyl fluoride, and 100 μg of lysozyme/ml, pH 7), and the cells were lysed by sonication in an Ultrasonicator (Misonix; Microson, New York, N.Y.). The lysed cells were centrifuged for 20 min at 5,000 × g and 4°C. Dehydrochlorinase activity in the supernatant was assayed by incubating 1 ml of cell extract (induced and uninduced E. coli cultures) with 5 μg of HCH isomer at 28°C with shaking. After regular time intervals, samples were withdrawn, extracted with hexane, and analyzed by GC as described above. The protein content of the cell extract was estimated by the method of Lowry et al. (13). Appropriate controls containing E. coli BL21 with pGEX-5X-3 were handled simultaneously.
Nucleotide sequence accession numbers.
The lin sequences of strain B90 have been deposited in GenBank under the following accession numbers: linX and linA1, AY150579; linA2, AY150580; linB, AY150581; linC, AY150582; and linD, AY150583.
RESULTS AND DISCUSSION
Degradation of HCH by S. paucimobilis B90.
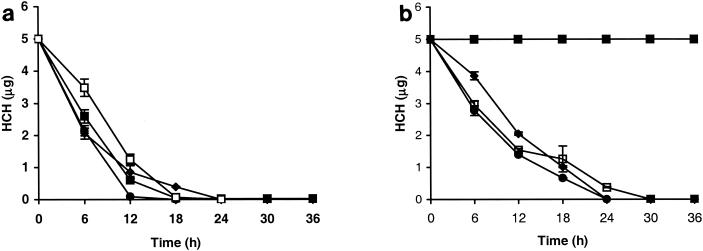
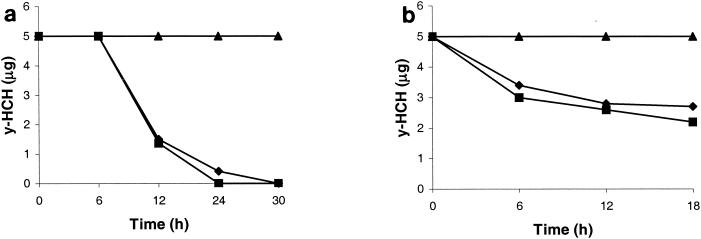
S. paucimobilis B90 was tested for its ability to degrade α-, β-, γ-, and δ-HCH separately at 5 μg/ml in SM medium containing 1% glucose. Strain B90 was found to degrade α-, γ-, and δ-HCH rapidly (Fig. 1a). β-HCH was also degraded, but it did not disappear completely from the medium (Fig. 1a). No α-, γ-, or δ-HCH was detectable in the medium after 24 h. No complete degradation took place at levels beyond 50 μg/ml (not shown). The rates of degradation of HCH isomers by S. paucimobilis strain B90 were slightly higher than those for S. paucimobilis UT26 (Fig. 1b). However, as expected, S. paucimobilis UT26 did not degrade β-HCH. Chromatograms obtained after GC analysis of the samples from incubations with α-, γ-, and δ-HCH revealed similar intermediates for S. paucimobilis B90 and S. paucimobilis UT26. The main types of intermediates corresponded to α-, γ-, and δ-PCCH, which was confirmed by PCCH standards produced after chemical conversion of each HCH isomer (α, γ, and δ; not shown). During β-HCH degradation, a very predominant peak appeared in GC chromatograms (at 6.5 min compared to β-HCH at 5 min) from samples taken after 36 and 72 h (not shown), which persisted in prolonged incubations but eventually disappeared. This intermediate had a different retention time than chemically synthesized β-PCCH. Further identification of this intermediate was outside the scope of the present work.
FIG. 1.
Degradation of HCH by S. paucimobilis B90 (a) and S. paucimobilis UT26 (b) in SM plus 1% glucose. An initial inoculum of 0.5 ml (108 cells/ml) was added to 50 ml of medium, and simultaneously, each HCH isomer was added separately (5 μg/ml). Samples were withdrawn periodically, extracted with hexane, and analyzed on a gas chromatograph equipped with an electron capture detector. ♦, α-HCH; ▪, β-HCH; •, γ-HCH; □, δ-HCH. The error bars indicate standard deviations.
Cloning of HCH dehydrochlorinase genes in E. coli.
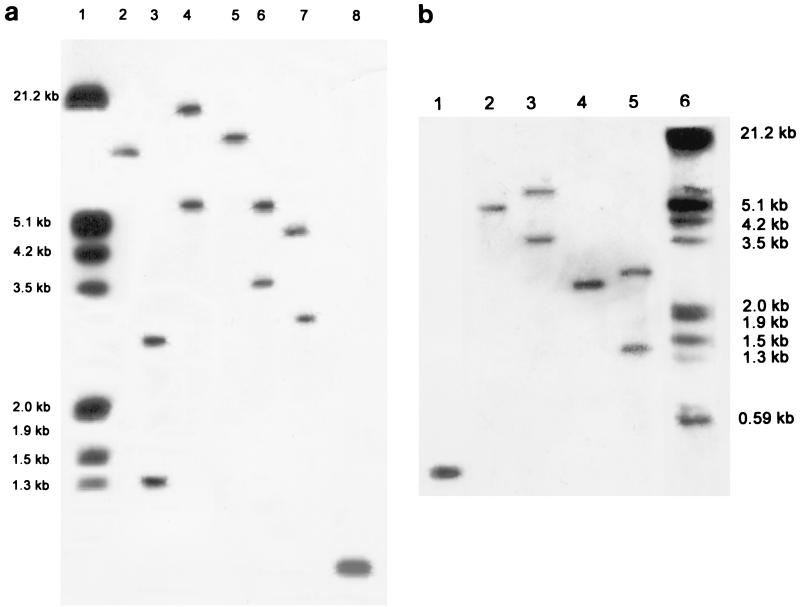
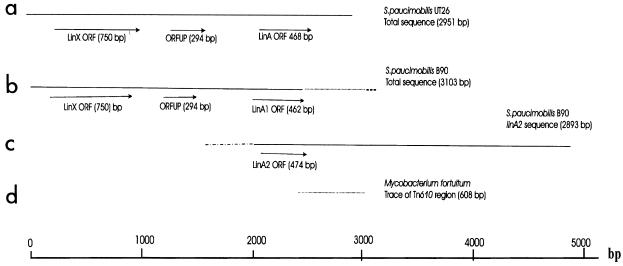
Genomic DNA of S. paucimobilis B90 produced strong positive hybridization signals when hybridized with a linA gene fragment of S. paucimobilis UT26 (Fig. 2). For a number of digests, two hybridizing bands were observed (Fig. 2b, lanes 3 and 5). The sequences of the two cloned S. paucimobilis B90 BclI fragments with sizes around 1.4 and 2.5 kb predicted two nonidentical linA-homologous genes (Table 3), named linA1 and linA2. The open reading frame (ORF) of the linA1 gene encoded a 154-amino-acid polypeptide with amino acids 88% identical to those of linA from strain UT26. The peptide predicted from the linA2 ORF encompassed 158 amino acids, with only the first two (extra) residues different from the linA gene product. The DNA sequence upstream and downstream of the linA1 gene differed from the linA2 region. Interestingly, the region upstream of linA1 showed strong similarity to the upstream region of linA in strain UT26 containing the linX gene and ORFUP (Fig. 3). Downstream of linA1, no homology to UT26 was found. Instead, the nucleotide sequence showed nucleotides 99% identical to the DNA sequence of Tn610 of Mycobacterium fortuitum (accession no. X536535). The 3′ end of the linA1 gene and the Tn610 sequence (only 608 nucleotides of the total nucleotide sequence of Tn610) appeared to overlap (Fig. 3). Although we did not isolate the Tn610 element completely from strain B90, hybridization of its genomic DNA with the DNA fragment from pLINA14 containing the Tn610-homologous DNA gave three hybridizing signals (data not shown), which indicated that there could be more than one copy of this element in the genome of S. paucimobilis B90. PCR amplification with primers based on the Tn610 sequence located downstream of the linA1 gene (primers 16 and 17 [Table 2]) resulted in a product of the expected size (655 bp) from S. paucimobilis B90 DNA but no amplification product with S. paucimobilis UT26 DNA (not shown). Also, genomic DNA of strain UT26 did not hybridize to the Tn610-like probe from plasmid pLIN14 in Southern hybridizations. This suggested that no Tn610-like element is present in strain UT26.
FIG. 2.
Southern blot hybridization of genomic DNA of S. paucimobilis B90 (a and b) and strain UT26 (b) with an [α-32P]dATP-labeled truncated fragment of linA. (a) Lane 1, lambda DNA digested with EcoRI and HindIII; lanes 2 to 7, B90 DNA digested with BamHI, BclI, BglII, EcoRI, HindIII, and SalI, respectively; lane 8, amplified linA1 (462 bp) as a positive control. (b) Lane 1, amplified linA1 (462 bp) as a positive control, lanes 2 to 5, genomic DNA digested with HindIII (UT26), HindIII (B90), BclI (UT26), and BclI (B90), respectively; lane 6, lambda DNA digested with EcoRI and HindIII.
TABLE 3.
Comparison of various HCH degradative genes in S. paucimobilis B90 and S. paucimobilis UT26
| Gene | Length (bp)
|
Peptide length (aa)a
|
Homology (%) | Function | Stabilityb
|
|||
|---|---|---|---|---|---|---|---|---|
| B90 | UT26 | B90 | UT26 | B90 | UT26 | |||
| linA1 | 462 | 468c | 154 | 156 | 96 | Dehydrochlorinase | + | − |
| linA2 | 474 | 468c | 158 | 100d | Dehydrochlorinase | + | − | |
| linB | 888 | 888 | 296 | 296 | 99 | Halidohydrolase | + | − |
| linC | 750 | 750 | 250 | 250 | 99 | Dehydrogenase | + | − |
| linD | 1,038 | 1,038 | 346 | 346 | 99 | Reductive dechlorinase | + | − |
| linE | 963 | 321 | Ring cleavage dioxygenase | ND | + | |||
| linR | 909 | 303 | Transcriptional regulator | ND | + | |||
aa, amino acids.
+, stable; −, unstable; ND, not detected.
linA gene of S. paucimobilis UT26
Percent identical nucleotides between linA and linA2
FIG. 3.
Schematic drawings of the regions from S. paucimobilis UT26 and B90 comprising the linA genes. (a) Reconstructed drawing from the published DNA sequences of linX (accession no. D23722) (20) and linA (accession no. D90355) (6) of S. paucimobilis UT26. (b) reconstructed drawings based on the sequences of linA1 and linX of S. paucimobilis B90. (c) The linA2 region. (d) Trace of Tn610 of M. fortuitum (accession no. X536535) (14a). Similar drawings (bold and dotted lines) indicate regions of DNA sequence homology between different genes or regions.
Conversely, the upstream sequence of the region of the linA2 gene did not show any homology to that of linA1 or linA of S. paucimobilis UT26, whereas its 3′ end completely overlapped with the DNA sequence downstream of linA of strain UT26 (Fig. 3). The present structure of the linA1 and linA2 regions in strain B90 suggests that the genetic organization of linX and linA in strain UT26 might have been the result of a recombination between the 5′ ends of (predecessor) linA1 and linA2 sequences. The few amino acid differences at the C-terminal end of LinA1 compared to those of LinA and LinA2 did not result in any obvious difference in dehydrochlorinase activity. Apparently, changes outside the catalytic dyad formed by His-73 and Asp-25 can be accommodated (34). Similar to linA of strain UT26 (6), the G+C contents of the linA1 and linA2 genes of S. paucimobilis B90 were lower than those of linX, linB, linC, and linD. Thus, as suggested earlier, the linA genes in strain B90 might have had a different origin than the other lin genes and perhaps were acquired by horizontal gene transfer (24).
Stability of the lindane degradation pathway.
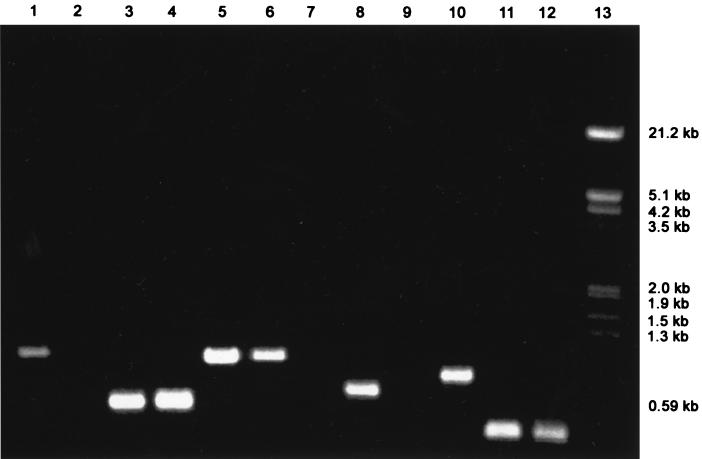
A few observations made in our laboratory suggest that the lindane degradation phenotype is more stable in strain B90 than in strain UT26. While linA1, linA2, linB, linC, and linD could always be amplified from colonies of S. paucimobilis B90 by the primers based on the corresponding lin genes from strain UT26 (Table 2), amplification could not be consistently achieved from colonies of S. paucimobilis UT26 (Fig. 4). When nearly 500 colonies of S. paucimobilis B90 were screened by GC analysis for degradation of HCH isomers, no lindane-negative mutants were detectable, whereas the same experiment with S. paucimobilis UT26 colonies resulted in 10 such mutants. The lack of the linA gene was confirmed in these UT26 mutants by PCR amplification with primers 1 and 2 and DNA-DNA hybridization with the α-32P-labeled linA as a probe (data not shown).
FIG. 4.
Detected instability of lin genes in strain UT26. Shown are products of PCR amplification with primers for linA, -B, -C, -D, and -X and the Tn610-like sequence on total DNA of S. paucimobilis B90 and UT26 isolated from individual clones. Lanes: 1 and 2, Tn610 detection in strain B90 (lane 1) and UT26 (lane 2); 3 and 4, linX in UT26 (lane 3) and B90 (lane 4); 5 and 6, linD in UT26 (lane 5) and B90 (lane 6); 7 and 8, linC in UT26 (lane7) and B90 (lane 8); 9 and 10, linB in UT26 (lane 9) and B90 (lane 10); 11 and 12, linA in UT26 (lane 11) and B90 (lane 12); 13, lambda DNA digested with EcoRI and HindIII.
Demonstration of functionality of both linA copies in strain B90.
By determining the disappearance of HCH from growing cultures of E. coli with plasmid pLINEA1 or pLINEA2, it was found that E. coli containing linA1 or linA2 degraded γ-HCH to γ-PCCH (Fig. 5a). No disappearance of γ-HCH was detected in control cultures of E. coli BL21 with pGEX-5X-3 alone. This demonstrated that the conversion was specific for the linA1 and linA2 inserts. Similar conversions of α- and δ-HCH to α- and δ-PCCH, respectively, were observed with E. coli BL21(pLINEA1) or BL21(pLINEA2) as well (not shown).
FIG. 5.
Degradation of γ-HCH by E. coli BL21(pLINEA1) or BL21(pGEX-5X-3). (a) Conversion of 5 μg of γ-HCH/ml by pLINEA1 and pLINEA2 in E. coli BL21 growing in LB medium. Samples were drawn periodically and analyzed by gas chromatography. ▴, control; ♦, pLINEA1; ▪, pLINEA2. (b) Degradation of γ-HCH by cell extracts of E. coli BL21(pLINEA1) expressing the linA1 gene. The cell extracts contained 5 mg of protein/ml, to which 5 μg of γ-HCH/ml was added. ▴, control; ♦, pLINEA1; ▪, pLINEA2.
Interestingly, the rate of degradation of γ-HCH was much lower in the beginning (up to 6 h) in E. coli BL21 containing pLINEA1 than that of pLINEA2 cultures (Fig. 5a). Whether this difference can be attributed to the differences between the amino acids in LinA1 and LinA2 has yet to be determined. β-HCH was not measurably converted by the E. coli linA1 or linA2 clone. The PCCH intermediates disappeared from growing cultures of E. coli BL21(pLINEA1) or BL21(pLINEA2), as well as in E. coli cell extracts upon prolonged incubation, which suggests their further dehydrochlorination.
Conversion of γ-HCH also took place in cell extracts of E. coli containing pLINEA1 or pLINEA2 (with PCCH as an intermediate), but not with E. coli BL21(pGEX-5X-3) (Fig. 5b). Again, only α-, γ-, and δ- but not β-HCH could be converted by the E. coli BL21(pLINEA1) or BL21(pLINEA2) cell extracts. The cell extracts prepared from IPTG (isopropyl-β-d-thiogalactopyranoside)-induced culture did not degrade α-, γ-, and δ-HCH, which reflected the fact that induction results in improper folding of proteins, making them inactive.
All of the results with respect to dehydrochlorinase activity and transient PCCH accumulation by LinA1 and LinA2 are consistent with those for LinA published by others (6, 23, 34). It might be that both linA copies in strain B90 result in slightly higher rates of HCH degradation than in strain UT26 (Fig. 1). Contrary to our expectations, the presence of both linA copies in strain B90 seemed not to be important for β-HCH conversion. Since degradation and activity toward α-, γ-, and δ-HCH did occur in E. coli BL21, this seems to rule out the possibility that the LinA protein is not correctly localized in E. coli. From these results, we conclude at this point that at least initial degradation of β-HCH proceeds through a different enzyme than LinA1 or LinA2.
Cloning of the linX, linB, linC, and linD genes of S. paucimobilis B90.
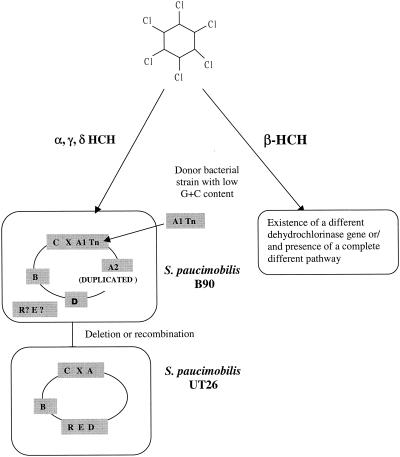
On the basis of presumed homology to the lin genes of strain UT26, the linX, linB, linC, and linD genes could also be identified and cloned from strain B90 by using primer sets listed in Table 2. These genes were highly similar to their counterparts in strain UT26 (Table 3). In contrast, no amplification of linE- and linR-like sequences in S. paucimobilis B90 could be obtained (data not shown). Hybridizations, restriction mapping, and DNA sequencing of cosmid clones containing linA, linB, linC, linX, and linD DNA fragments revealed that no single cosmid contained a DNA insert encompassing all of the isolated lin genes from strain B90 (Table 4). The linA1 copy associated with the Tn610-like fragment and linX and linC (Fig. 3), whereas the linA2 gene and the other lin genes (linB and linD) were not associated and were present at different positions on the B90 genome. The strong homology of the other lin ORFs of strain B90 with those of strain UT26 (Table 3) strongly suggests that they carry out the same functions in strain B90. Therefore, it is very likely that the lindane degradation pathway in strain B90 is similar to that of strain UT26, at least up to the step of hydroquinone (Fig. 6). Since we did not obtain amplification or hybridization of linE or linR, the ring cleavage oxygenase and regulatory element for the lower pathway might be different from that of strain UT26 (24).
TABLE 4.
Hybridization mapping of cosmid clones with linA, linB, linC, linD, linX, and Tn610 as probes
| DNA probe | Hybridization with cosmid clonea:
|
|||||
|---|---|---|---|---|---|---|
| 3 | 5 | 6 | 10 | 23 | 27 | |
| linA1 | − | − | + | + | + | + |
| linA2 | − | − | + | + | + | + |
| linB | − | + | − | − | − | − |
| linC | − | − | + | − | + | + |
| linD | + | − | − | − | − | − |
| linX | − | − | + | − | + | + |
| Tn610 | − | − | + | − | + | + |
+, hybridization; −, no hybridization.
FIG. 6.
Possible evolutionary origin of HCH degradative genes and pathways in S. paucimobilis B90 and S. paucimobilis UT26.
It is definitely intriguing and hopeful that at least three different aerobic lindane-degrading bacteria isolated at different positions on the globe (S. paucimobilis B90 from India, S. paucimobilis UT26 from Japan [6], and S. paucimobilis from France [33]) carry very similar if not identical genetic information for lindane degradation. Such microorganisms may lead to the spontaneous disappearance of HCH residues in the environment or might be used as a basis for achieving targeted bioremediation of HCH isomers.
Acknowledgments
We thank Y. Nagata, Department of Biotechnology, University of Tokyo, for providing the plasmids and S. paucimobilis UT26. Thanks are also due to R. Eichenlaub and K. H. Gartemann for valuable suggestions and help. We also thank Kirsten Lawlor for her help in providing the linE probe and her valuable suggestions for the detection of linE and linR.
Part of this work was supported by grants under the Indo-Swiss Collaboration in Biotechnology.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann, A., P. Walet, P. Wijnen, W. de Bruin, J. L. M. Huntjens, W. Roelofsen, and A. J. B. Zehnder. 1988. Biodegradation of alpha- and beta-hexachlorocyclohexane in a soil slurry under different redox conditions. Appl. Environ. Microbiol. 54:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhuyan, S., B. Sreedharan, T. K. Adhya, and N. Sethunathan. 1993. Enhanced biodegradation of γ-hexachlorocyclohexane (γ-HCH) in HCH (commercial) acclimatized flooded soil: factors affecting its development and persistence. Pestic. Sci. 38:49-55. [Google Scholar]
- 4.Cerkvenik, V., D. Z. Doganoc, and J. Jan. 2000. Evidence of some trace elements, organochlorines pesticides and PCBs in Slovenian cow's milk. Food Technol. Biotechnol. 38:155-160. [Google Scholar]
- 5.Imai, R., Y. Nagata, K. Senoo, H. Wada, M. Fukuda, M. Takagi, and K. Yano. 1989. Dehydrochlorination of γ-hexachlorocyclohexane (γ-BHC) by γ-BHC-assimilating Pseudomonas paucimobilis. Agric. Biol. Chem. 53:2015-2017. [Google Scholar]
- 6.Imai, R., Y. Nagata, M. Fukuda, M. Takagi, and K. Yano. 1991. Molecular cloning of a Pseudomonas paucimobilis gene encoding a 17-kilodalton polypeptide that eliminates HCl molecules from γ-hexachlorocyclohexane. J. Bacteriol. 173:6811-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen, A. A., and S. A. Slorach. 1991. Chemical contaminants in human milk. CRC Press, Inc., Boca Raton, Fla.
- 8.Johri, A. K., M. Dua, D. Tuteja, R. Saxena, D. M. Saxena, and R. Lal. 1996. Genetic manipulations of microorganisms for the degradation of hexachlorocyclohexane. FEMS Microbiol. Rev. 19:69-84. [DOI] [PubMed] [Google Scholar]
- 9.Johri, A. K., M. Dua, D. Tuteja, R. Saxena, D. M. Saxena, and R. Lal. 1998. Degradation of alpha, beta, gamma, and delta-hexachlorocyclohexane by Sphingomonas paucimobilis. Biotechnol. Lett. 20:885-887. [Google Scholar]
- 10.Lal, R., and D. M. Saxena. 1982. Accumulation, metabolism and toxic effects of organochlorine insecticides on microorganisms. Microbiol. Rev. 46:95-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lal, R., S. Lal, P. S. Dhanraj, and D. M. Saxena. 1995. Manipulation of catabolic genes for the degradation and detoxification of xenobiotics. Adv. Appl. Microbiol. 41:55-95. [DOI] [PubMed] [Google Scholar]
- 12.Li, Y. F., R. W. Macdonald, L. M. Jantunen, T. Harner, T. F. Bidleman, and W. M. Strachan. 2002. The transport of beta-hexachlorocyclohexane to the western Arctic Ocean: a contrast to alpha-HCH. Sci. Total Environ. 27:229-246. [DOI] [PubMed] [Google Scholar]
- 13.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin Phenol Reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 14.MacRae, I. C., K. Raghu, and E. M. Bautista. 1969. Anaerobic degradation of the insecticide lindane by Clostridium sp. Nature (London) 221:859-860. [DOI] [PubMed] [Google Scholar]
- 14a.Martin, C., J. Timm, J. Rauzier, R. Gomez-Luz, J. Davies, and B. Gicquel. 1990. Transposition of an antibiotic resistance element in mycobacteria. Nature 345:739-743. [DOI] [PubMed] [Google Scholar]
- 15.Matsumura, F., H. J. Benzet, and K. C. Patil. 1976. Factors affecting microbial metabolism of γ-BHC. J. Pestic. Sci. 1:3-8. [Google Scholar]
- 16.Miyauchi, K., S. Suh, Y. Nagata, and M. Takagi. 1998. Cloning and sequencing of a 2,5-dichlorohydroquinone reductive dehalogenase gene which is involved in the degradation of γ-hexachlorocyclohexane in Sphingomonas paucimobilis. J. Bacteriol. 180:1354-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyauchi, K., Y. Adachi, Y. Nagata, and M. Takagi. 1999. Cloning and sequencing of a novel meta-cleavage dioxygenase gene whose product is involved in degradation of γ-hexachlorocyclohexane in Sphingomonas paucimobilis. J. Bacteriol. 181:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyauchi, K., H. S. Lee, M. Fukuda, M. Takagi, and Y. Nagata. 2002. Cloning and characterization of linR, involved in regulation of the downstream pathway for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 68:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagasawa, S., R. Kikuchi, Y. Nagata, M. Takagi, and M. Matsuo. 1993. Aerobic mineralization of γ-HCH by Pseudomonas paucimobilis UT 26. Chemosphere 26:1719-1728. [Google Scholar]
- 20.Nagata, Y., R. Ohtomo, K. Miyauchi, M. Fukuda, K. Yano, and M. Takagi. 1994. Cloning and sequencing of a 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase gene involved in the degradation of γ-hexachlorocyclohexane in Pseudomonas paucimobilis. J. Bacteriol. 176:3117-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagata, Y., K. Miyauchi, J. Damborsky, K. Manova, A. Ansorgova, and M. Takagi. 1997. Purification and characterization of a haloalkane dehalogenase of a new substrate class from a γ-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT 26. Appl. Environ. Microbiol. 63:3707-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagata, Y., T. Hatta, R. Imai, K. Kimbara, M. Fukuda, K. Yano, and M. Takagi. 1993. Purification and characterization of γ-hexachlorocyclohexane (γ-HCH) dehydrochlorinase (LinA) from Pseudomonas paucimobilis. Biosci. Biotechnol. Biochem. 57:1582-1583. [DOI] [PubMed] [Google Scholar]
- 23.Nagata, Y., T. Nariya, R. Ohtomo, M. Fukuda, K. Yano, and M. Takagi. 1993. Cloning and sequencing of a dehalogenase gene encoding an enzyme with hydrolase activity involved in the degradation of γ-hexachlorocyclohexane in Pseudomonas paucimobilis. J. Bacteriol. 175:6403-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagata, Y., K. Miyauchi, and M. Takagi. 1999. Complete analysis of genes and enzymes for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. J. Ind. Microbiol. Biotechnol. 23:380-390. [DOI] [PubMed] [Google Scholar]
- 25.Nagata, Y., A. Futamura, K. Miyauchi, and M. Takagi. 1999. Two different types of dehalogenases, LinA and LinB, involved in γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26 are localized in the periplasmic space without molecular processing. J. Bacteriol. 181:5409-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nigam, U., and K. J. Siddqui. 2001. Organochlorine insecticide residues in dairy milk samples collected in Lucknow, India. Bull. Environ. Contam. Toxicol. 66:678-682. [DOI] [PubMed] [Google Scholar]
- 27.Noren, K., and D. Meironyte. 2000. Organochlorine and organobromine contaminants in Swedish human milk in perspective of past 20-30 years. Chemosphere. 44:1111-1123. [DOI] [PubMed] [Google Scholar]
- 28.Ohisa, N., and M. Yamaguchi. 1978. Gamma BHC degradation accompanied by the growth of Clostridium rectum isolated from paddy field soil. Agric. Biol. Chem. 42:1819-1823. [Google Scholar]
- 29.Sahu, S. K., K. K. Patnaik, M. Sharmila, and N. Sethunathan. 1990. Degradation of alpha-, beta-, and gamma-hexachlorocyclohexane by a soil bacterium under aerobic conditions. Appl. Environ. Microbiol. 56:3620-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Senoo, K., and H. Wada. 1989. Isolation and identification of an aerobic γ-HCH-decomposing bacterium from soil. Soil Sci. Plant Nutr. 35:79-87. [Google Scholar]
- 32.Simonich, S. L., and R. A. Hites. 1995. Global distribution of persistent organochlorine compounds. Science 269:1851-1854. [DOI] [PubMed] [Google Scholar]
- 33.Thomas, J. C., F. Berger, M. Jacquier, D. Bernikkon, F. Baud-Grasset, N. Truffaul, P. Normand, T. M. Vogel, and P. Simonet. 1996. Isolation and characterization of a novel γ-hexachlorocyclohexane-degrading bacterium. J. Bacteriol. 178:6049-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trantirek, L., K. Hynkova, Y. Nagata, A. Murzin, A. Ansorgova, V. Sklenar, and J. Damborsky. 2001. Reaction mechanism and stereochemistry of γ-hexachlorocyclohexane dehydrochlorinase LinA. J. Biol. Chem. 276:7734-7740. [DOI] [PubMed] [Google Scholar]
- 35.Wada, H., K. Senoo, and Y. Takai. 1989. Rapid degradation of γ-HCH in upland soil after multiple applications. Soil Sci. Plant Nutr. 35:71-77. [Google Scholar]
- 36.Windholz, M., S. Budavari, L. Y. Stroumtsos, and M. N. Fertig (ed.). 1976. The Merck index, 9th ed., p. 719-720. Merck & Co., Inc., Rahway, N.J.