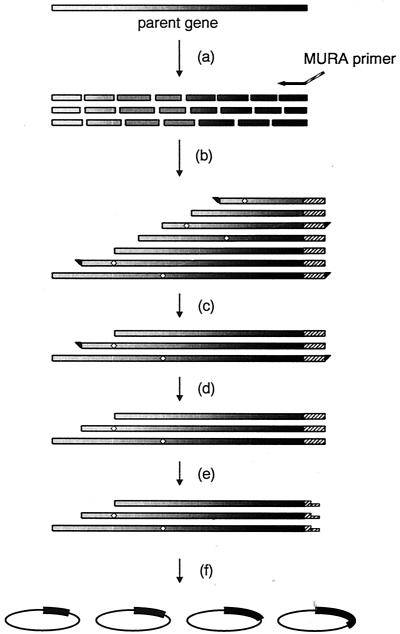
FIG. 1.
Schematic overview of the construction of a DNA-shuffled and truncated enzyme library by the MURA method. The sequences of the MURA primer can be chosen on any desired site of the parent gene. This figure represents a procedure using a 3′-complementary MURA primer, and thus N-terminal-truncated and shuffled variants of PlaA are constructed. The steps shown are random fragmentation of the parent gene pool by DNase I (a), unidirectional reassembly with the MURA primer (b), separation of the fragments of interest by preparative agarose gel electrophoresis (c), formation of blunt ends by S1 nuclease or T4 DNA polymerase on both termini (d), sticky-end formation by a restriction enzyme on one terminus (e), and ligation into an expression vector (f). The order of steps c, d, and e can be altered to d, e, and c without affecting the efficiency of library construction. The diamonds on the bars representing genes indicate possible mutations during shuffled and unidirectional reassembly PCR.