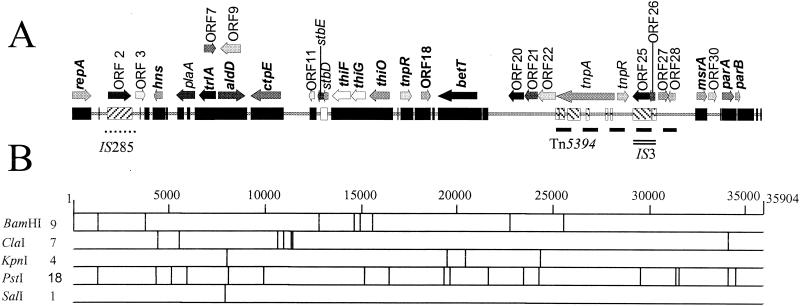
FIG. 4.
Physical and restriction map of 35,904-bp plasmid pEP36 isolated from E. pyrifoliae strain Ep1/96. (A) Genetic map of pEP36. Solid black bar (▪) indicates nucleotide homology to plasmid pEA29 isolated from E. amylovora strain Ea88. The forward-hatched box (▨), open box (□), and reverse-hatched box (▧) indicated regions of pEP36 with nucleotide similarities to the Y. pestis strain CO92 genome, E. amylovora plasmid pEA29 isolated from Rubus species, and an S. flexneri SHI-2 pathogenicity island, respectively; all nucleotide similarities reported are greater than 80%. The location of insertion elements IS285 and IS3 are shown by a dotted line (••••) and double line (=), respectively. The position of the transposon Tn5394 is designated by a dashed line (---). Potential ORFs greater than 375 nt and putative genes inferred from sequence data or with matches in GenBank are indicated by directional arrows. ORFs conserved between pEP36 and E. amylovora plasmid pEA29 are indicated in bold lettering. A key to the ORFs is found in Table 2. (B) Restriction map of pEP36. Numbers to the right of the restriction enzyme are indicative of the number of restriction sites found for each enzyme on the plasmid. Only nonmethylated ClaI sties are reported.