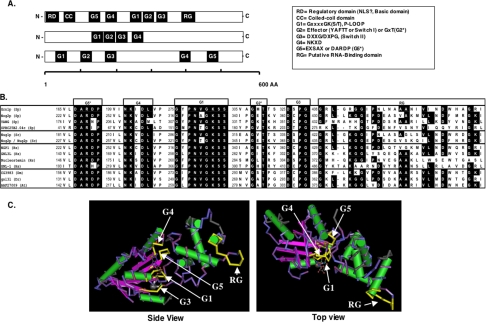
Figure 1.
Grn1p is member of a unique family of HSR1_MMR1-nucleolar GTPases with a highly conserved circularly permuted “G”-domain. (A) Schematic representation of three types of G-proteins illustrating the relative positions of the motifs that make up the G-domain. Top bar: GTPases with a circular permutation of the classic G-domain; middle bar: a small group of GTPases that belong to the Nog-subfamily; bottom bar represents the “classic” G-proteins as exemplified by the Ras, EF-2, and heterotrimeric G-protein families. (B) Alignment of the above circularly permuted G-domain showing individual motifs G5*, G4, G1, G2*, G3, and the putative RNA-binding domain (RG). Motifs G5* and G2* correspond to G5-like and G2-like, respectively (see text). Representatives were chosen from a broad selection of eukaryotes: S. pombe (Sp), S. cerevisiae (Sc), human (Hs), Drosophila melanogaster (Dm), Danio rerio (Dm), and Arabidopsis thaliana (At). Identical residues are shaded black (conservative substitutions are not indicated). Numbers to the left of each motif indicate the beginning of the amino acid motif. (C) 3D representation of the highly conserved circularly permuted GTPases based on the structure of Bacillus subtilis Ylqf GTPase as the consensus for the HSR1_MMR1 GTP-binding domain from the 3D-structure Entrez database, MMDB (http://www.ncbi.nlm.nih.gov/Structure/MMDB/mmdb.shtml). Images were visualized and recorded using NCBI's Cn3D4.1 software. G-motifs and the RG-domain are indicated with arrows. Flat arrows represent β-sheets, whereas cylinders represent α-helices.