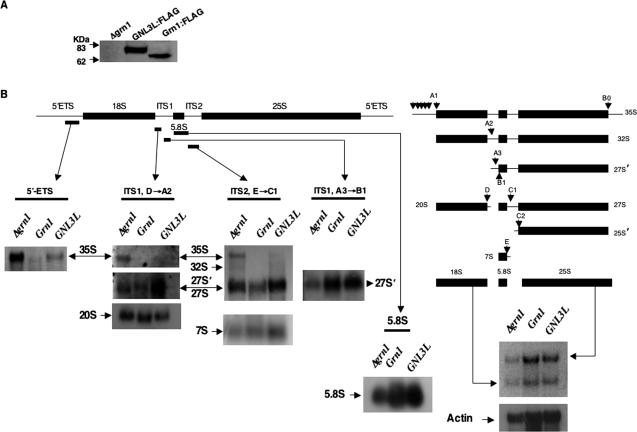
Figure 3.
Effect of Grn1p and GNL3L on processing of 35S pre-rRNA species. (A) Grn1:FLAG (YNB859) and GNL3L:FLAG (YNB858) were tested for genomically expressed FLAG-tagged Grn1p and GNL3L by Western analysis and probing with anti-FLAG. The null mutant (YNB484) was used as control. (B) Pre-rRNA and mature rRNA species were detected in the above strains by Northern hybridizational analysis. DNA probes specific for 5′ ETS, 5.8S, ITS1 or ITS2 are indicated by bars under the respective flanks. The rRNA processing pathway was adapted from Good et al. (1997). Downward pointed arrows indicate relative positions of processing sites.