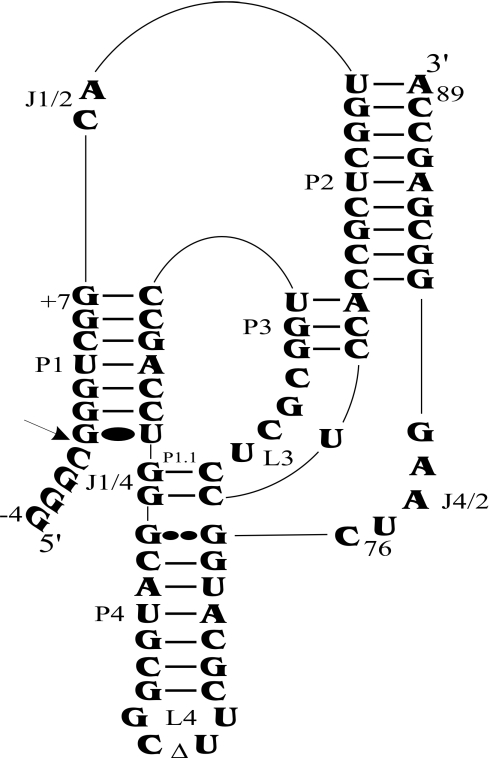
Figure 1.
Secondary structure of antigenomic self-cleaving HDV RNA. The numbering system used is that of Shih and Been (2). The triangle in the L4 loop indicates the P4 deletion (as compared with the natural variants). The homopurine base pair at the top of the P4 stem is represented by two large dots (G••G), while the Wobble base pair is represented by a single large dot (G•U). The arrow indicates the cleavage site.