Abstract
Cell surface mannose 6-phosphate/insulin-like growth factor II receptors (M6P/IGF2R) bind and target exogenous insulin-like growth factor II (IGF2) to the prelysosomes where it is degraded. Loss of heterozygosity (LOH) for M6P/IGF2R is found in cancers, with mutational inactivation of the remaining allele. We exploited the normal allele-specific differential methylation of the M6P/IGF2R intron 2 CpG island to rapidly evaluate potential LOH in ovarian cancers, since every normal individual is informative. To this end, we developed a method for bisulfite modification of genomic DNA in 96-well format that allows for rapid methylation profiling. We identified ovarian cancers with M6P/IGF2R LOH, but unexpectedly also found frequent abnormal acquisition of methylation on the paternally inherited allele at intron 2. These results demonstrate the utility of our high-throughput method of bisulfite modification for analysis of large sample numbers. They further show that the methylation status of the intron 2 CpG island may be a useful indicator of LOH and biomarker of disease.
INTRODUCTION
The mannose 6-phosphate/insulin-like growth factor II receptor (M6P/IGF2R) encodes a multifunctional protein that is involved in lysosomal sorting, latent TGFβ activation and binding the potent mitogen, insulin-like growth factor II (IGF2). IGF2 binding to the M6P/IGF2R causes IGF2 to be internalized and degraded in the prelysosomal compartment (1). Loss of M6P/IGF2R function leads to increased extracellular IGF2 and decreased activated TGFβ. These molecular effects increase cell proliferation, decrease apoptosis, and enhance tumor invasion and metastasis, suggesting that IGF2R functions as a tumor suppressor (2). This is supported by observations indicating that loss of heterozygosity (LOH) is frequent in breast, lung and liver cancers, with mutational inactivation of the remaining allele (3–6).
Mouse M6p/Igf2r is subject to genomic imprinting (7,8), a form of epigenetic regulation that results in parent of origin dependent monoallelic expression. M6p/Igf2r has two differentially methylated CpG islands. The first (region 1) encompasses the promoter and is methylated on the repressed paternally inherited allele, whereas the intron 2 CpG island (region 2) is methylated on the expressed maternally inherited allele (9). Region 2 methylation is established during oogenesis and constitutes the inherited imprint mark, whereas region 1 is methylated postfertilization (9). An M6p/Igf2r antisense transcript is produced from within region 2 in mice on the paternally inherited chromosome and this is required for imprinted expression of M6p/Igf2r. For all tissues thus far examined, humans do not exhibit imprinted expression of M6P/IGF2R, with biallelic expression evident throughout the primate lineage (10). Furthermore, an antisense transcript in humans has not been reported. However, similar to mice, humans do exhibit maternal methylation of orthologous region 2 (11). It is unclear whether the differential methylation of region 2 in humans is functionally relevant, or a vestige of the evolutionary loss of imprinted expression.
Previous microarray analysis of 58 advanced stage serous epithelial ovarian cancers (12) indicated that the average transcription of the M6P/IGF2R is decreased in most cancers compared with that observed for normal ovarian surface epithelium (N = 3; Affymetrix U133A GeneChips; 858.7 versus 2064.2, respectively, P = 0.04, t-test, Welch-corrected). This difference in expression might be due to LOH. We were therefore interested in generating an assay to rapidly screen for potential M6P/IGF2R LOH that exploits the differentially methylated status of region 2. Such a technique would improve upon genetic polymorphism-based analysis in that every sample is ‘heterozygous’ by virtue of the allele-specific differential methylation of this region.
Sodium bisulfite converts unmethylated cytosines to uracil, while methylated cytosines are unaffected (13). Bisulfite treatment of DNA is presently a widely used tool in conducting DNA methylation analysis, and most studies employ either commercially available kits or variations of the method pioneered by Frommer et al. (13), and more recently optimized by Grunau et al. (14). The hands-on manipulations required using these methods limit the number of samples that can be simultaneously treated (in our hands, ∼20) and this potentially influences the consistency between preparations. In the present study, we modified the smaller scale bisulfite treatment protocol, (hereafter referred to as SSBM, for single sample bisulfite modification) to develop an assay for high-throughput bisulfite modification (HTBM) taking advantage of a 96-well format. We validated this assay by analyzing the differentially methylated region (DMR) associated with the imprinted maternally expressed gene 3 (MEG3) (15). We then used HTBM to prepare samples for analysis of M6P/IGF2R region 2 in ovarian and breast cancers. Surprisingly, we found a very high frequency of hypermethylation in both types of cancer. We confirmed suspected LOH based on this methylation profile in ovarian cancers using genetic polymorphism analysis.
MATERIALS AND METHODS
Tissue samples and nucleic acid purification
Normal human genomic DNA used for validation and comparison of the HTBM and SSBM methods was purchased from Promega (Madison, WI). CpGenome Universal Methylated DNA (Chemicon International; Temecula, CA) was used as a control. Genomic DNA was prepared using PUREGENE Reagents (Gentra Systems; Minneapolis, MN). All tissues were obtained with patient consent. The Duke University Medical Center Institutional Review Board approved this study.
Bisulfite modification of genomic DNA
High-throughput bisulfite modification
Based on experimental parameters published by Grunau et al. (14), we adjusted the reaction components proportionately to accommodate a 96-well format. Incubation steps were performed in a heated lid thermocycler. DNA (3 ng to 2 µg in a 10 µl volume) was denatured with 1.1 µl freshly prepared 3 M NaOH (VWR; Westchester, PA) for 20 min at 42°C, then mixed with 120 µl sodium bisulfite solution (saturated sodium bisulfite, 10 mM hydroquinone, pH 5.0). The bisulfite solution was prepared as follows: 10.8 g sodium bisulfite (Sigma-Aldrich; St Louis, MO) was brought to 16 ml final volume in water and inverted gently to mix. We added 2.6 ml freshly prepared 3 M NaOH followed by 1.0 ml hydroquinone solution [prepared by dissolving 0.22 g hydroquinone (Sigma-Aldrich) in 10 ml water]. Undissolved bisulfite will remain in the solution since it is saturated; we therefore filtered the solution prior to use with a 0.45µ filter. The samples in bisulfite solution were incubated for 4 h at 55°C.
An aliquot of 100 µl nuclease-free water was then added to each well followed by transfer to a Montage PCR96 96-well filtration plate (Millipore, Billerica, MA). The size exclusion filtration matrix in these plates prevents flow-through of fragments >200 bp. All remaining steps of the protocol were performed in the Montage PCR96 filtration plate using a vacuum manifold (Millipore MultiScreen Vacuum Manifold) and an in-house vacuum source. The solution was drawn through the filtration matrix by vacuum (20–25 inches Hg) until the wells were visibly empty of solution, typically requiring about 5 min. The DNA was desalted with 175 µl nuclease-free H2O three times followed by desulfonation with 175 µl of 0.1 M NaOH (with immediate vacuum filtration) and a final wash step with 175 µl H2O. The DNA was recovered in 50 µl of 10 mM Tris/HCl (pH 8.0) by using a plate shaker to release the DNA from the filtration matrix for 10 min (Vortex Genie 2, setting ∼5) followed by transfer to individual tubes and storage at −20°C.
Single sample bisulfite modification (SSBM)
SSBM followed the method described previously (14,15) except that the DNA was resuspended in 50 µl 10 mM Tris/HCl, pH 8.0 and stored at −20°C.
Methylation-specific PCR (MS-PCR)
MS-PCR and bisulfite sequencing for the DLK1/MEG3 DMR were performed as described previously (15) without multiplexing the reactions.
The primers for MS-PCR of M6P/IGF2R region 2 were MF 5′-CGG TTT TTT GCG GTT TTT CGT C-3′; MR 5′-AAC CCA ACC GCA CGA CCA C-3′; UF 5′-GGT TTT TTG TGG TTT TTT GTT GGT GTT G-3′; and UR 5′-AAT AAC CCA ACC ACA CAA CCA CAC TAA CA-3′. The primers for M6P/IGF2R region 1 were MF 5′-GTT GTC GTC GAG TTT AGT CGA GTC-3′; MR 5′-GCG CGA CTA CTA AAA CCG-3′; UF 5′-GTT GTT GTT GAG TTT AGT TGA GTT GTG-3′; and UR 5′-CTA ACA ACA CAC AAC TAC TAA AAA CCA C-3′. All primers were obtained from Sigma-Genosys (The Woodlands, TX).
Bisulfite-treated DNA (30–60 ng) was amplified in a 25 µl reaction volume containing 1× reaction buffer, 3 mM MgCl2, 0.2 mM each dNTP, 2.5 U Platinum Taq DNA Polymerase (Invitrogen; Carlsbad, CA), and 0.4 µM each forward and reverse M or U primer. Cycling conditions for M6P/IGF2R region 2 MS-PCR were 3 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 69°C and 30 s at 72°C and final extension of 5 min at 72°C. The cycling conditions for M6P/IGF2R region 1 were 3 min at 94°C followed by 40 cycles of 30 s at 94°C, 45 s at 62°C and 45 s at 72°C and final 5 min extension at 72°C.
Bisulfite sequencing
Bisulfite-treated genomic DNA was PCR amplified (3 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 55°C and 30 s at 72°C with a 5 min final extension at 72°C) with primers specific for bisulfite converted MEG3 DMR as described previously (15) in a 25 µl reaction volume containing 3 µl of each titration of the bisulfite-treated DNA template, 3 mM MgCl2, 0.2 mM dNTPs and 0.4 µM each primer. The 235 bp PCR products were resolved on a 2.5% agarose gel, purified using Sigma GenElute spin columns (Sigma; St Louis, MO) and cycle sequenced (15) (Thermo Sequenase Radiolabeled Terminator Cycle Sequencing kit; Amersham Biosciences, Piscataway, NJ). Thermocycler conditions were 35 cycles of 30 s at 95°C, 30 s at 55°C and 60 s at 72°C. The sequencing products were resolved on a 5% denaturing polyacrylamide sequencing gel followed by exposure at −80°C to radiographic film with an intensifying screen.
Analysis of cloned amplicons from HTBM-treated genomic DNA
PCR amplicons were generated using 40 ng bisulfite modified DNA with primers specific for M6P/IGF2R region 2: F1, 5′-TGA ATT TGG AGT TTG GGG TTT GTA GTG TTG GGA GGT-3′ and R1, 5′-CTA ACA AAT TCA AAA ACC AAA CCC CCA AC-3′. Because of the very limited DNA from normal OSEs 1–5, 1 µl was used as template in a 10 µl reaction volume. All other PCRs were 25 µl total volume. PCR conditions were 94°C for 3 min followed by 40 cycles of 94°C for 30 s, 66°C for 30 s and 72°C for 30 s with a final 5 min extension at 72°C. The amplicons were resolved on 2% agarose gels, purified using the Qiagen MinElute PCR Cleanup kit (Valencia, CA), ligated into pGEMT-Easy vectors (Promega; Madison, WI) and plasmids were transformed into competent JM109 Escherichia coli (Promega; Madison, WI) followed by plating to LB agar with 100 µg/ml ampicillin (TEKNOVA; Hollister, CA). Whole cell PCR was used to amplify plasmids from individual colonies using SP6 and T7 primers (94°C for 5 min, then 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 30 s, final 5 min extension at 72°C). The amplicons were resolved on agarose gels, purified, and sequenced using the F1 primer.
Laser capture microdissection and LOH analysis
Seven micron sections were cut from frozen Tissue-Tek® cryomold blocks (Sakura; Torrance, CA) and briefly stained with hematoxylin and eosin prior to capture of normal and malignant cells. Slides were dehydrated using a series of graded ethanol washes. Laser capture was performed using the Veritas Microdissection System (Arcturus Bioscience, Inc.; Mountain View, CA). DNA from normal and malignant cells was prepared using PUREGENE reagents (Gentra Systems; Minneapolis, MN) followed by genotyping with primers that flank polymorphic sites located in the M6P/IGF2R intron 2 DMR (rs8191722) (forward: 5′-CTG GCA TGT TGG GGA CAG GT-3′; reverse: 5′-GAA GCG GGA AGG GAG TGA CC-3′) and/or an A/G SNP located at position 2286 of the M6P/IGF2R transcript (NM_000876) in exon 16 (forward: 5′-GTG ACT CCT CAC GTC GCT CAC G-3′; reverse: 5′-CAC AGG CAT GAG TAT CCT CAG G-3′). The PCR amplicons were purified using Sigma GenElute spin columns (Sigma-Aldrich; Saint Louis, MO) followed by nucleotide sequencing using the primer 5′-CTG CCT GGC CCT TGT CTG G-3′ for intron 2 and primer 5′-GGG TCT GAG TAA TGC GAA GC-3′ for exon 16. The sequencing reactions were resolved on 5% denaturing polyacrylamide gels, dried and analyzed using Phosphorimager analysis (Storm 840 Scanner and ImageQuant Software; GE Healthcare Piscataway, NJ). An allelic imbalance in the malignant cells >4:1 was considered positive for LOH.
RESULTS
Comparison of bisulfite modification methods and validation of HTBM
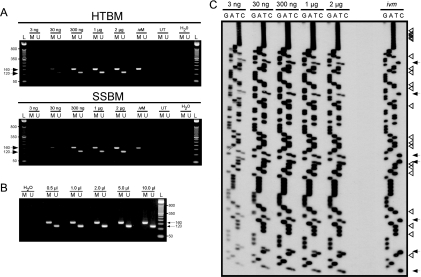
We treated increasing amounts of human genomic DNA using both methods and assessed the results using MS-PCR and bisulfite sequencing of the MEG3 DMR on human chromosome 14q32.2 (15) (Figure 1). Universally methylated (in vitro methylated; ivM) and untreated (UT) human genomic DNA were included as controls. As shown in Figure 1A, the primers specific for methylated (M) and unmethylated (U) DNA produce 160 and 120 bp amplicons, respectively. An aliquot of 2 µl of each DNA sample was used as template for MS-PCR from the 50 µl used to recover the modified DNA. MS-PCR amplicons were detectable for both the U and M primers using both methods, even from 3 ng of treated DNA.
Figure 1.
Comparison of HTBM and SSBM and validation of HTBM. (A) Increasing amounts of human genomic DNA were modified using HTBM and SSBM followed by MS-PCR of the MEG3 DMR on human chromosome 14q. An aliquot of 500 ng CpGenome Universal Methylated DNA (ivM) was also modified using each method and used as a control. Both methylated (M, 160 bp) and unmethylated (U, 120 bp) DNA products were detected using bisulfite-treated samples. L, 50 bp ladder; ivM, Universal Methylated DNA; UT, untreated genomic DNA; H20, water control. (B) MS-PCR of the MEG3 DMR using increasing amounts of the 300 ng input genomic DNA treated using the HTBM protocol. (C) Validation of conversion efficiency of HTBM by bisulfite sequencing of the MEG3 DMR. The indicated amounts of normal human genomic DNA and 500 ng of the Universal Methylated DNA (ivm) were prepared using HTBM, amplified using primers specific for the MEG3 DMR that do not anneal to regions containing CpG dinucleotides, and manually sequenced. Cytosines in 5′-CpG-3′ context in the original unconverted sequence are indicated by black arrows, while non-CpG cytosines are indicated by the open arrowheads.
Using 0.5–10 µl of the 300 ng input HTBM sample as template, we also determined the amount of modified DNA required to produce optimal yields of MEG3 MS-PCR amplicons (Figure 1B). The results indicate that 0.5 µl is sufficient, although increasing the template produced higher yields and did not appear to compromise specificity.
Bisulfite sequencing is regarded as a comprehensive method for determining methylation status since it provides information for each CpG dinucleotide as well as a visual representation of the conversion efficiency for non-CpG (unmethylated) cytosines. In order to assess conversion efficiency of the HTBM method, we performed bisulfite sequencing of the MEG3 DMR (15) using DNA treated in increasing amounts by HTBM (Figure 1C). The control in vitro methylated template showed complete methylation of each CpG. Because this region is differentially methylated, we observed equivalent amounts of unmethylated and methylated cytosines at each CpG position in the sequence for the normal DNA (arrows). Importantly, each non-CpG cytosine was fully converted by HTBM (arrowheads). Surprisingly, we obtained good quality sequence from the amplicon generated from the 3 ng sample, indicating that degradation was negligible during the HTBM procedure.
Analysis of M6P/IGF2R methylation status in normal and malignant tissues
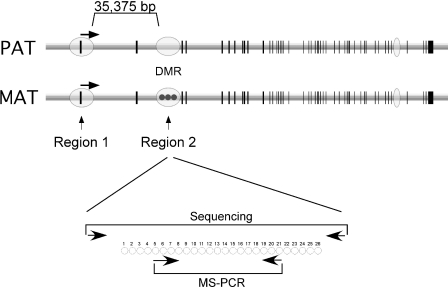
We used HTBM to prepare genomic DNA from cancers and normal lymphocytes to determine the utility of using the differential methylation of M6P/IGF2R region 2 as an indicator for potential LOH. The M6P/IGF2R consists of 48 exons spanning ∼137.5 kb (Figure 2) with three associated CpG islands: one encompassing the promoter that is normally unmethylated (region 1), one in intron 2 that exhibits differential methylation (region 2) and one in intron 44 whose methylation status is unknown.
Figure 2.
M6P/IGF2R genomic structure. The M6P/IGF2R locus consists of 48 exons (vertical lines) and is associated with three CpG islands (ovals). Region 1 is the promoter CpG island, while region 2 is a DMR located within intron 2. Region 2 is characterized by maternal (MAT) allele-specific methylation, while the paternal allele (PAT) is unmethylated. The direction of transcription is indicated. The relative location of primers used for sequencing and for methylation-specific PCR is shown below. Numbered circles represent CpG dinucleotides present within the region (not to scale) and analyzed in the present study.
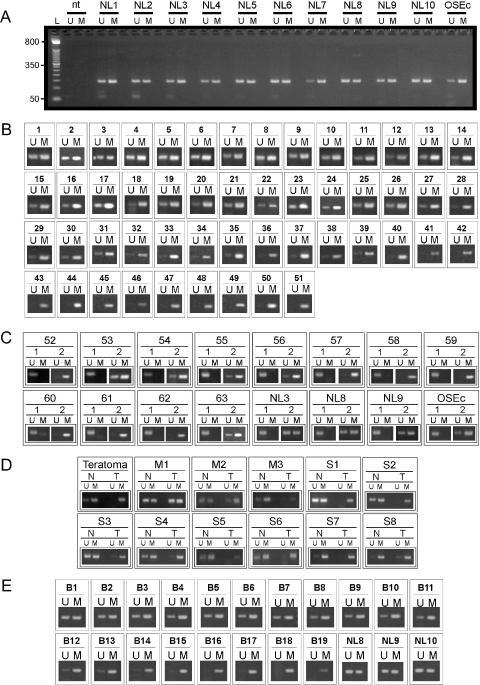
We treated 300 ng genomic DNA from each specimen, recovered the DNA in 50 µl and used 2 µl of this as template in 25 µl MS-PCRs. Most normal lymphocyte samples gave a roughly equal distribution of unmethylated and methylated amplicons by MS-PCR (Figure 3A). One showed slightly more methylated amplicons for region 2 (NL7), indicating that there may be inter-individual variability in the degree of methylation present in this region. This was also noted for a normal ovarian surface epithelium sample that had undergone several passages in culture (OSEc). MS-PCR analysis of DNA from over 60 serous epithelial ovarian tumors showed the majority exhibit an increased yield of methylated amplicons for region 2, with some producing only methylated amplicons (Figure 3B).
Figure 3.
Methylation-specific PCR of M6P/IGF2R region 2. (A) MS-PCR of region 2 using HTBM-treated peripheral blood lymphocytes (NL) and a primary culture of normal ovarian surface epithelium (OSEc). L, 50 bp ladder; U, primers used specific for unmethylated DNA which produce a 139 bp amplicon; M, primers used specific for methylated DNA which produce a 137 bp amplicon; nt, H2O control. (B) HTBM DNA from primary serous epithelial ovarian cancers was treated using HTBM and analyzed by MS-PCR as described in (A). (C) HTBM DNA from primary serous epithelial ovarian cancers was subjected to MS-PCR with primers for the M6P/IGF2R promoter CpG island (region 1, ‘1’) and to region 2 (‘2’). (D) HTBM DNA from matched lymphocytes (N) and ovarian malignancies (M, mucinous; S, serous) was subjected to MS-PCR for M6P/IGF2R region 2 as described above. (E) M6P/IGF2R MS-PCR of region 2 using HTBM DNA from primary breast cancers (B) and normal lymphocytes (NL).
To determine whether the apparently increased methylation at region 2 of M6P/IGF2R is associated with methylation spreading from an aberrantly methylated promoter CpG island (region 1), we analyzed 12 tumors by MS-PCR for regions 1 and 2 along with several normal lymphocyte samples (NL3, NL8 and NL9) and OSEc (Figure 3C). In all lymphocytes and in the OSEc sample, both methylated and unmethylated amplicons were obtained for region 2 while only unmethylated amplicons were obtained for region 1. For 11 of the tumors with increased region 2 methylation, region 1 was completely unmethylated. In one case (number 60), methylation was indeed detected at region 1, suggesting the possibility of a regional methylation defect in this individual. Nevertheless, these results indicate that methylation spreading from the promoter is not associated with the frequent increase in methylated amplicons detected for region 2.
To establish whether the methylation changes observed by MS-PCR were specific to the malignancies, we analyzed HTBM tumor and matched lymphocyte DNA by MS-PCR (Figure 3D). Complete methylation was observed in an ovarian teratoma for region 2, while the matched lymphocytes yielded both unmethylated and methylated amplicons by MS-PCR. The absence of the unmethylated (paternally derived) amplicon in the teratoma is indicative of the maternal derivation of the genetic material in these types of neoplasms. For mucinous ovarian cancers M1 and M2, the methylation profiles did not differ substantially between tumor and lymphocytes, while for M3, a stronger signal was present in both tissues for methylated region 2. Matched lymphocytes from four of eight serous ovarian tumors had relatively normal differential methylation (S1–S4), while lymphocytes from S5–S8 appeared to be hypermethylated. Every one of these serous cancers exhibited increased methylated amplicons. We cannot rule out at this point that the increased methylation detected in lymphocytes is not from circulating tumor DNA. This seems unlikely since the methylation is very prominent in each case, and such a result would require large amounts of tumor DNA in the peripheral circulation to mask the signal from the lymphocytes. These results suggest that altered methylation of region 2 may represent a global attribute that is present from early development in some individuals, while in others the increase in methylation is specific to the malignancy.
The frequent increased M6P/IGF2R region 2 methylation detected in ovarian cancer prompted us to evaluate whether this is also evident in other types of cancer. We analyzed 19 infiltrating ductal breast carcinoma DNA samples using HTBM followed by MS-PCR for region 2. As shown in Figure 3E, almost half of these tumors (numbers B12-B19) also exhibited increased or complete methylation of this region.
Verification of M6P/IGF2R MS-PCR results by sequencing of cloned alleles
In order to verify the methylation status of M6P/IGF2R region 2 detected by MS-PCR, we cloned PCR amplicons generated using primers that anneal to non-CpG containing sequence and that flank the region analyzed by MS-PCR (see Figure 2). Clones from normal lymphocyte NL11 fell into two prominent groups, as 7 of 13 clones sequenced were mostly unmethylated at individual CpG dinucleotides within this region and the remaining six were mostly methylated (Figure 4A). For samples NL3 and NL12, we were able to distinguish the two alleles because of a single nucleotide polymorphism (G/T; rs8191722) present within the sequenced region. The G allele of NL3 was mostly methylated (96%), while the T allele was less methylated (54%). For NL12, both the G and T alleles exhibited higher methylation (95 and 79%, respectively). These results indicate that there is variability in the methylation status of this region in these peripheral blood cells.
Figure 4.
Analysis of M6P/IGF2R region 2 methylation status by cloning and sequencing individual alleles from HTBM-treated DNA. Each row of squares corresponds to one individual clone; columns represent each cytosine in CpG context within the sequenced region. Filled box, methylated; unfilled, unmethylated. (A) Normal peripheral blood lymphocytes (NL) from women without evident malignancy. NL3 and NL12 were heterozygous for a T/G SNP within the sequenced region allowing the individual parental alleles to be identified. (B) Sequenced clones from normal ovarian surface epithelium (OSE). OSEs denotes samples that were obtained by gently scraping the surface epithelium from the ovary during surgical oophorectomy; OSEc are normal OSE cells that underwent several passages in tissue culture prior to DNA extraction and analysis. (C) Methylation status of alleles cloned from ovarian malignancies that exhibited differential methylation (3) and hypermethylation (39, 52, 43) by MS-PCR. (D) Methylation status of cloned alleles from ovarian malignancies heterozygous for one of two polymorphisms (a T/G or a C/T SNP) present within the sequenced region, allowing for discrimination between parental alleles; all of these specimens exhibited hypermethylation by MS-PCR.
We also analyzed five normal ovarian surface epithelium samples (Figure 4B, OSEs1-5) that were obtained by gentle intra-operative scraping of the surface epithelium prior to oophorectomy. For all OSEs samples we observed a differential methylation pattern, in that the clones for each could be grouped into mostly methylated or mostly unmethylated categories. We also examined the normal OSE cells that had been expanded by several passages in tissue culture prior to harvest and DNA purification (OSEc, see Figure 3A and C for MS-PCR result). In this case, there was a more random distribution of CpG methylation in the clones (Figure 4B). These results show that normal ovarian surface epithelium exhibits a predominant differentially methylated pattern for this region, while culture of normal OSE is associated with region 2 hypermethylation.
We next analyzed a series of tumors that were determined to exhibit different types of methylation patterns by MS-PCR. In Figure 3B, tumor 3 was scored as differentially methylated by MS-PCR, and the sequence of the cloned alleles in Figure 4C supports this finding. By MS-PCR, tumor 39 appeared more methylated and 52 and 43 as predominantly methylated, and this is again in agreement with the sequencing results.
M6P/IGF2R region 2 is hypermethylated in ovarian cancer
In order to distinguish between the possibilities that this region is truly hypermethylated versus the alternative explanation that the observed methylation profile is due to loss of heterozygosity, we genotyped samples for the SNP mentioned above, and in the process identified an additional SNP for several samples located three bp upstream from rs8191722 (a CpG-altering C/T polymorphism; rs677882). We sequenced individual clones from seven tumors that were scored as hypermethylated or fully methylated by MS-PCR (Figure 3B and C) and that were informative for one of these SNPs. We determined that, in each case, both alleles were indeed present and they both exhibited methylation. In some instances, the pattern of methylation was suggestive of parental origin. For example, the T allele in tumor 55, and the G alleles in tumors 47 and 59 were likely derived from the paternally inherited chromosome, since they exhibit unmethylated CpGs in a higher proportion than the opposite allele. Importantly, these results indicate either that LOH is not present in these tumors, or if it is present, paternal allele hypermethylation at region 2 of M6P/IGF2R occurred prior to LOH.
M6P/IGF2R loss of heterozygosity in ovarian cancer
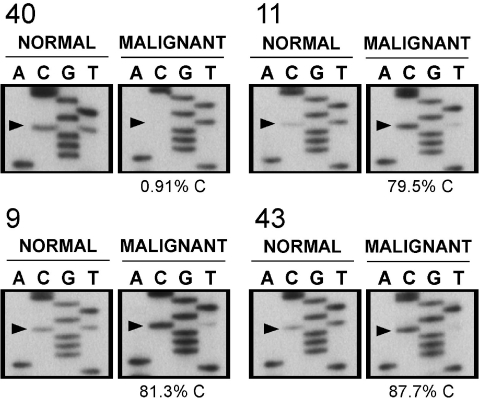
In order to determine whether LOH was indeed present in ovarian malignancies, we analyzed a polymorphism at mRNA position 2286 in exon 16 of M6P/IGF2R (5) in four heterozygotes using laser capture microdissected (LCM) normal and malignant cell populations (Figure 5). We considered an allelic distribution ratio that exceeds 4:1 (>80% loss) to be indicative of LOH; three of the four cancers analyzed met this criterion. The remaining specimen exibited a 3.9:1 allelic distribution, marginal for LOH. In each case, the MS-PCR results (Figure 3B and C) also showed an increased methylation profile (9,11,40), or complete methylation (43), supporting our conclusion that MS-PCR of region 2 is a useful method to screen for potential LOH.
Figure 5.
Loss of heterozygosity analysis in laser capture microdissected ovarian malignancies. Normal stroma and malignant cell populations were enriched using laser capture microdissection for cancer specimens heterozygous for a SNP in exon 16 of M6P/IGF2R to assess for potential allelic loss. Peripheral blood lymphocytes were available for analysis in one case (number 40). Nucleotide sequencing was followed by PhosphorImager analysis to quantify each allele, shown as the percent of the C allele present for this C/T polymorphism. Loss of heterozygosity, defined by an allelic imbalance >4:1 in the malignant cells, is evident in three of the four samples analyzed.
DISCUSSION
Alterations in DNA methylation are among the most common molecular alterations in human malignancies. Detection of aberrant methylation associated with cancer-related genes is a promising approach to augment cancer diagnosis and improve treatment options. Bisulfite modification is prerequisite for most modern techniques aimed at detecting changes in methylation, but has been limited by throughput capacity. In this study, we developed and verified a high-throughput method for bisulfite modification and have used it to analyze aberrant methylation associated with the M6P/IGF2R in epithelial ovarian and breast cancer.
HTBM offers several practical advantages over SSBM. First, the 96-well plate format simplifies sample handling and minimizes potential loss. HTBM eliminates the DNA precipitation step required in the SSBM protocol, insuring good recovery, and the need to resuspend the DNA pellet, avoiding unnecessary shearing. No mineral oil overlay is required, and complete treatment takes about 6 h. Because of decreased handling, it is possible to perform parallel reactions in multiple plates. This format also opens the possibility of utilizing automation.
We found that as little as 3 ng of input genomic DNA in both methods allowed for recovery of amplicons by MS-PCR and for bisulfite sequencing. The apparent high degree of sensitivity may be a result of the use of commercially available, high quality genomic DNA for these experiments. In this regard, however, we also successfully used HTBM for MS-PCR and bisulfite sequencing utilizing minimal DNA from scraped OSE (Figure 4B) and LCM samples (data not shown). We noted that optimal detection of products from these specimens occurred within the first several weeks post-HTBM with storage at −20°C. When 300 ng or more genomic DNA was treated using HTBM, we were able to obtain PCR amplicons for up to 6 months.
M6P/IGF2R is not imprinted in brain, kidney, heart, liver, lung or placenta (10). Imprinting in other human tissues has not been systematically investigated, but recent studies indicate a maternal effect in the transmission of type 1 diabetes (16), suggestive of the potential for tissue-specific imprinting. It is curious that in each case of distorted methylation, hypermethylation was always observed rather than hypomethylation. Biased loss of the paternal allele might indicate that the gene is imprinted in these tissues. However, if M6P/IGF2R is imprinted in humans as it is in mice, the maternal allele is expected to be active. As such, it is unclear why the silent allele of this putative tumor suppressor gene would be lost, unless the driving force is another imprinted locus in proximity to M6P/IGF2R. The most parsimonious explanation for our findings is that region 2 is vulnerable to acquisition of hypermethylation. This could result from a cellular milieu that directs aberrant methylation to target regions, and the differentially methylated status of region 2 may create a methylation ‘sink’ for the paternal allele. This effect, if correct, is limited to this CpG island since the promoter CpG island was very infrequently found to exhibit methylation. Alternatively, hypermethylation may influence a previously unappreciated function for region 2 in the transcription of M6P/IGF2R. Our microarray data (12) support the former hypothesis, in that there is no statistical correlation between M6P/IGF2R expression and hypermethylation at region 2 (data not shown). It is presently unclear whether there is a functional link between hypermethylation of region 2 and LOH.
M6P/IGF2R region 2 methylation status in ovarian and breast tumors may be useful as a biomarker of disease. We demonstrated not only that region 2 hypermethylation is an indicator of potential LOH, but also that region 2 was very frequently methylated on both parental alleles. The finding that LOH was present in three of four serous epithelial ovarian cancers exhibiting region 2 hypermethylation indicates that the analysis of this region should prove useful for the study of other cancers in which LOH is prominent. The other aspect of this study, the preparation of genomic DNA using HTBM for methylation analysis, will prove beneficial for any epigenetic study focused on analysis of DNA methylation in a large number of specimens.
Acknowledgments
The authors thank Nancy Glover in the Department of Surgery, Duke University for preparing slides for LCM. The authors also thank Christophe Grunau for advice and Randy Jirtle for helpful discussions. Funding to S.K.M. from the Ovarian Cancer Research Fund and the Marsha Rivkin Center for Ovarian Cancer Research supported this work. Funding to pay the Open Access publication charges for this article was provided by the Gail Parkins Ovarian Awareness Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pollak M.N., Schernhammer E.S., Hankinson S.E. Insulin-like growth factors and neoplasia. Nat. Rev. Cancer. 2004;4:505–518. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 2.De Souza A.T., Yamada T., Mills J.J., Jirtle R.L. Imprinted genes in liver carcinogenesis. FASEB J. 1997;11:60–67. doi: 10.1096/fasebj.11.1.9034167. [DOI] [PubMed] [Google Scholar]
- 3.Hankins G.R., De Souza A.T., Bentley R.C., Patel M.R., Marks J.R., Iglehart J.D., Jirtle R.L. M6P/IGF2 receptor: a candidate breast tumor suppressor gene. Oncogene. 1996;12:2003–2009. [PubMed] [Google Scholar]
- 4.Kong F.M., Anscher M.S., Washington M.K., Killian J.K., Jirtle R.L. M6P/IGF2R is mutated in squamous cell carcinoma of the lung. Oncogene. 2000;19:1572–1578. doi: 10.1038/sj.onc.1203437. [DOI] [PubMed] [Google Scholar]
- 5.Oka Y., Waterland R.A., Killian J.K., Nolan C.M., Jang H.S., Tohara K., Sakaguchi S., Yao T., Iwashita A., Yata Y., et al. M6P/IGF2R tumor suppressor gene mutated in hepatocellular carcinomas in Japan. Hepatology. 2002;35:1153–1163. doi: 10.1053/jhep.2002.32669. [DOI] [PubMed] [Google Scholar]
- 6.Leboulleux S., Gaston V., Boulle N., Le Bouc Y., Gicquel C. Loss of heterozygosity at the mannose 6-phosphate/insulin-like growth factor 2 receptor locus: a frequent but late event in adrenocortical tumorigenesis. Eur. J. Endocrinol. 2001;144:163–168. doi: 10.1530/eje.0.1440163. [DOI] [PubMed] [Google Scholar]
- 7.Barlow D.P., Stoger R., Herrmann B.G., Saito K., Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 8.Kalscheuer V.M., Mariman E.C., Schepens M.T., Rehder H., Ropers H.H. The insulin-like growth factor type-2 receptor gene is imprinted in the mouse but not in humans. Nat. Genet. 1993;5:74–78. doi: 10.1038/ng0993-74. [DOI] [PubMed] [Google Scholar]
- 9.Stoger R., Kubicka P., Liu C.G., Kafri T., Razin A., Cedar H., Barlow D.P. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 10.Killian J.K., Nolan C.M., Wylie A.A., Li T., Vu T.H., Hoffman A.R., Jirtle R.L. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum. Mol. Genet. 2001;10:1721–1728. doi: 10.1093/hmg/10.17.1721. [DOI] [PubMed] [Google Scholar]
- 11.Smrzka O.W., Fae I., Stoger R., Kurzbauer R., Fischer G.F., Henn T., Weith A., Barlow D.P. Conservation of a maternal-specific methylation signal at the human IGF2R locus. Hum. Mol. Genet. 1995;4:1945–1952. doi: 10.1093/hmg/4.10.1945. [DOI] [PubMed] [Google Scholar]
- 12.Berchuck A., Iversen E.S., Lancaster J.M., Pittman J., Luo J., Lee P., Murphy S., Dressman H.K., Febbo P.G., West M., et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin. Cancer Res. 2005;11:3686–3696. doi: 10.1158/1078-0432.CCR-04-2398. [DOI] [PubMed] [Google Scholar]
- 13.Frommer M., McDonald L.E., Millar D.S., Collis C.M., Watt F., Grigg G.W., Molloy P.L., Paul C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grunau C., Clark S.J., Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65–65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy S.K., Wylie A.A., Coveler K.J., Cotter P.D., Papenhausen P.R., Sutton V.R., Shaffer L.G., Jirtle R.L. Epigenetic detection of human chromosome 14 uniparental disomy. Hum. Mutat. 2003;22:92–97. doi: 10.1002/humu.10237. [DOI] [PubMed] [Google Scholar]
- 16.McCann J.A., Xu Y.Q., Frechette R., Guazzarotti L., Polychronakos C. The insulin-like growth factor-II receptor gene is associated with type 1 diabetes: evidence of a maternal effect. J. Clin. Endocrinol. Metab. 2004;89:5700–5706. doi: 10.1210/jc.2004-0553. [DOI] [PubMed] [Google Scholar]