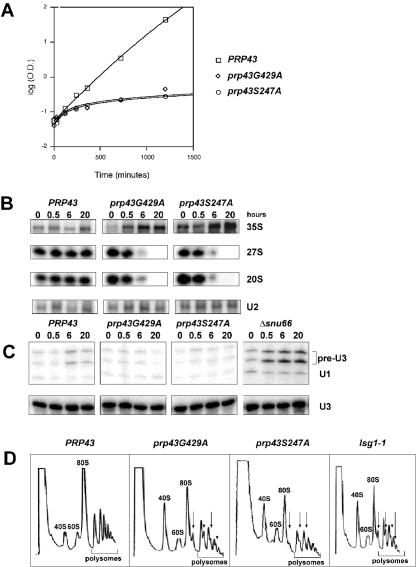
FIG. 3.
Growth defects in prp43 mutants correlate to rRNA processing defects but not pre-U3 splicing defects. (A) Mutant prp43 strains are growth impaired at 16°C. Growth curves of PRP43 (open box), prp43G429A (open diamond), and prp43S247A (open circle) after the shift to the nonpermissive temperature (16°C) are shown. (B) Pre-rRNA processing defects are detected in prp43 mutant strains. Shown is Northern blot analysis of total RNA from strains in panel A over a 20-h time course. Note the accumulation of 35S pre-rRNA and the depletion of 27S and 20S pre-rRNA over time. (C) Mutant prp43 strains do not exhibit pre-mRNA splicing defects in pre-U3A or pre-U3B snoRNA introns. RNA from strains grown as described above (A) and a Δsnu66 control were subjected to primer extension analysis using oligonucleotide primers designed to detect U3 pre-snoRNA, U3 snoRNA, and U1 snRNA (loading control). (D) Mutant prp43 strains are depleted for ribosomal subunits at nonpermissive temperatures. Sucrose gradient polysome profiles are shown for PRP43, prp43S247A, prp43G429A, and the lsg1-1 control. The direction of the increasing sedimentation coefficient is from left to right. Ribosomal subunits, polysomes, and half-mers are noted.