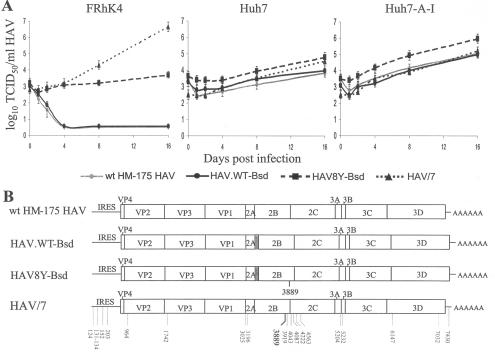
FIG. 5.
Growth of wt HAV in different cell lines. (A) One-step growth curve analysis of wt HAV recombinant viruses containing the Bsd selectable marker (HAV8Y-Bsd and HAV.WT-Bsd), wt HAV isolated from human stools (wt HM-175 HAV), and cell culture-adapted HAV (HAV/7) was done in monkey kidney (FRhK-4) cells, human hepatoma (Huh7) cells, and interferon-cured (Huh7-A-I) cells. Viral titers were assessed in Huh7-A-I cells using the ELISA endpoint titration assay as indicated in Fig. 4C. (B) Schematic representation of nucleotide sequences. Full-length nucleotide sequences of HAV8Y-Bsd, HAV.WT-Bsd, wt HM-175 HAV, and HAV/7 RNA extracted from the last time point sample of the growth curve in Huh7-A-I cells were obtained by RT-PCR. Sequences that differ from the wt HM-175 HAV are indicated with the nucleotide position under the viral genomes. The main cell culture-adapting mutation at nt 3889 is indicated in boldface. Viral genes, the IRES, and the 3′-end poly(A) tail are indicated. The bsd gene clone in HAV8Y-Bsd and HAV.WT-Bsd is shown as a gray box.