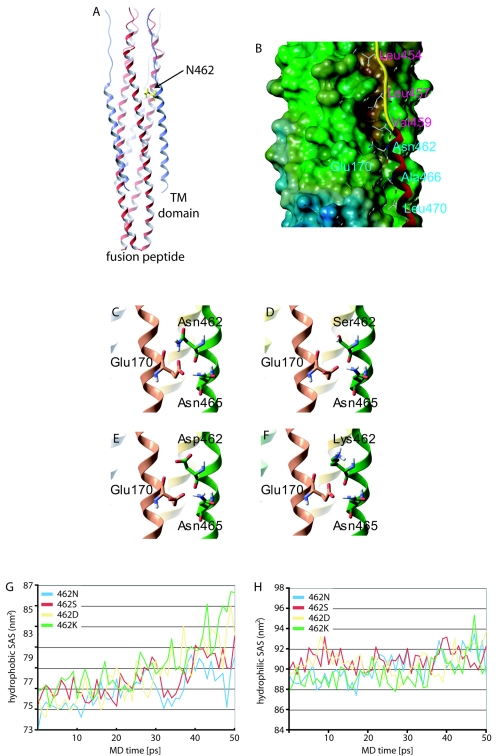
FIG. 2.
Localization of residue 462 in the final MV F core structure. (A) Ribbon model of the MV 6-HB; N termini of HR-B domains are facing up. For clarity, residue 462 is highlighted in only one of the three HR-B domains. TM, transmembrane. (B) Surface model of the MV 6-HB, shown with only one HR-B ribbon for clarity. Residues V459, L457, and L454 are predicted to interact with a hydrophobic groove in the HR-A trimer. HR-A residue E170 is predicted to engage in hydrogen bonding with residues N462 and N465. (C to F) Enlarged ribbon models of HR-A and HR-B highlighting the interaction described above (C). Dynamic structural modeling predicts disruption of the hydrogen bonding, destabilizing the interaction for either of the resistant mutants based on distance (D), charge (E), or steric hindrance (F). (G and H) MD simulations predict greater destabilization of the mutant 6-HBs compared to the wild type. The mutants increase the peptides' hydrophobic exposure to water through the course of the simulation, indicating greater dissociation (G). Hydrophobic solvent-accessible surface (SAS) areas are shown for 50-ps MD simulation. Hydrophilic exposures to water were similar for the four structures, with average hydrophilic SAS areas over the last 10 picoseconds of simulation between 91.1 nm2 and 92.0 nm2 (H).