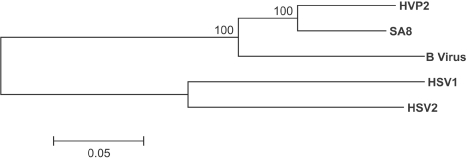
FIG. 1.
Phylogenetic relationship among simplexviruses. Whole genome alignments of HVP-2, SA8, B virus, HSV-1, and HSV-2 were compiled and compared using Mega v3.0 with ClustalW, with a gap-opening penalty of 20 and a gap extension penalty of 1 for both the pairwise and multiple parameters. Bootstrap analysis was performed using neighbor joining and the Kimura 2-parameter model with 500 replicates. Gaps or missing data were treated with complete deletion of the sites for the analysis. The resulting bootstrap values are indicated on the dendrogram.