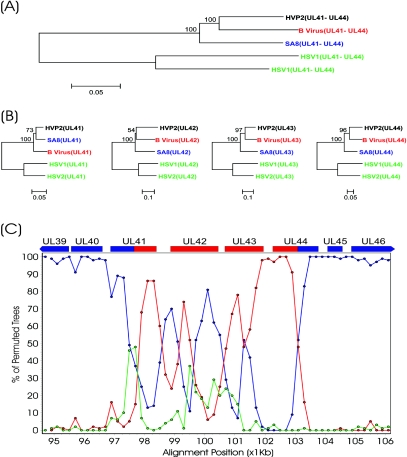
Fig. 3.
Putative recombination event in the UL41/UL44 region of the HVP-2 genome. DNA-based bootstrap analysis of the UL41/UL44 region (A) and comparison of the individual proteins from this region (B) are shown. Alignments were compiled and compared using Mega v3.0 with ClustalW. Bootstrap analysis was performed using neighbor joining and the Kimura 2-parameter (DNA) or Poisson correction (protein) models with 500 replicates. The resulting bootstrap values are indicated on the dendrograms. (C) SimPlot analysis of the UL41/UL44 region. Bootscan analysis was conducted on the whole genome alignment illustrated in Fig. 1 using a window setting of 1,000 bp and a step value of 200 bp. The program did not perform bootstrap analysis in a given window if the percentage of gaps in the alignment exceeded 50% (gap stripping set to 50%). Using HVP-2 as the query sequence, the comparison with SA8 is plotted in blue, B virus in red, and the HSV-1/HSV-2 consensus in green.