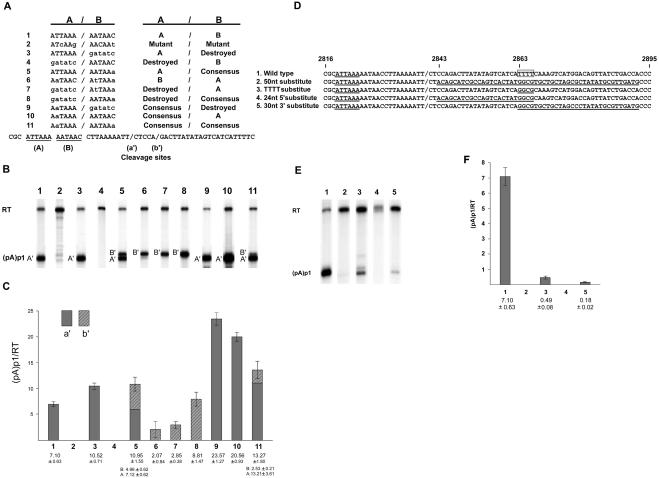
FIG. 2.
Characterization of the core hexanucleotide and downstream element required for polyadenylation at (pA)p1. (A) The (pA)p1 extended core polyadenylation motif, indicating the A and B components (line 1), as well as all mutations of these components (lines 2 to 11), as described in the text. The two cleavage sites (designated a′ and b′), as determined by anchored RT-PCR as previously described (10), are indicated in the sequence at the bottom. (B) RNase protection assays, performed as previously described (7, 12) using probe 4 (see Fig. 1), of RNA generated in COS7 cells following transfection of wild-type or mutant plasmids as characterized in panel A. Bands representing RNAs that either read through (RT) or are polyadenylated at (pA)p1 are designated to the left of the gel. (C) Quantification, using a Molecular Imager FX and Quantity One version 4.2.2 image software (Bio-Rad, Hercules, CA), of the RNase protections shown in panel B. Data from at least three experiments are presented as the ratio of transcripts terminated at (pA)p1 relative to those reading through. The filled and hatched portions of the bars represent (pA)p1 polyadenylated RNAs that are cleaved at sites a′ and b′, respectively. Error bars indicate standard deviations. (D) Nucleotide sequence of B19 from nt 2816 to 2895, which encompasses the downstream element required for polyadenylation at (pA)p1, for the wild type and the mutants described in the text. The essential ATTAAA hexanucleotide starting at nt 2819 for each is underlined, the a′ cleavage site is indicated by a slash, and mutated sequences downstream of the cleavage site that have been changed are also underlined. The 4-nt poly(T) motif is indicated within an open square. (E) RNase protection assays, performed as previously described (7, 12) using probe 4 (see Fig. 1), of RNA generated in COS7 cells following transfection of wild-type or mutant plasmids. Bands representing RNAs that either read through or are polyadenylated at (pA)p1 are designated to the left of the gel. (F) Quantification, as in panel C, of RNase protection assays as shown in panel E. Data from at least three experiments are presented as the ratio of transcripts terminated at (pA)p1 relative to those reading through. All RNAs generated by these mutants were cleaved at site a′.