FIG. 4.
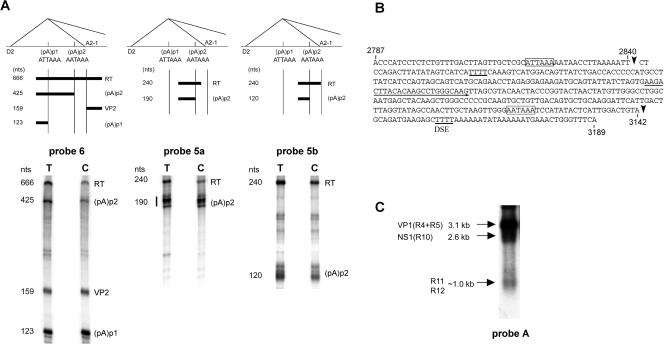
Approximately 10% of B19 P6-generated RNA polyadenylated in the center of the genome uses a second site, (pA)p2. (A) RNase protection assays showing the usage of (pA)p2. The top shows the locations of the probes used for protection assays (probes 6, 5a, and 5b, left to right) in relation to (pA)p1 and (pAp)2, the intron donor D2 and acceptor A2-1, and the bands predicted to be protected by RNAs polyadenylated at the two sites. RNase protection assays, performed as previously described (7, 12), of RNA generated in COS7 cells following transfection of pC1NS(−), using the three probes, are shown underneath each relevant diagram. The identities of the bands are labeled and correlate to the diagram above. RT, readthrough. (B) Nucleotide sequence of B19 from nt 2787 to 3189, showing the locations of the main (pA)p1 cleavage site (a′) at nt 2840 and of the cleavage site for (pA)p2 at nt 3142 (arrows). The (pA)p1 hexanucleotide motif identified in Fig. 2 and the putative consensus AATAAA signal of (pA)p2 are indicated by open boxes. Note that there are four T residues 36 nt downstream of this site, which could function as a downstream element (DSE). The (pA)p2 cleavage site was identified from RNA generated following both pC1NS(−) transfection of COS7 cells and B19 infection of erythropoietin-treated UT-7/EpoS1 cells (6), by anchored RT-PCR as previously described (10), using a forward primer which is indicated by an underline and was the same for both cell types. (C). Northern analysis, using probe A, of RNA generated following pC1NS(−) transfection of COS7 cells. The identities of the RNA transcripts are shown on the left. The probe, which lies downstream of (pA)p1, hybridizes with VP1-encoding R4 and R5; RNA R10, which could encode NS1; R11, which encodes the 7.5-kDa protein; and R12, from which no other protein product has been described.