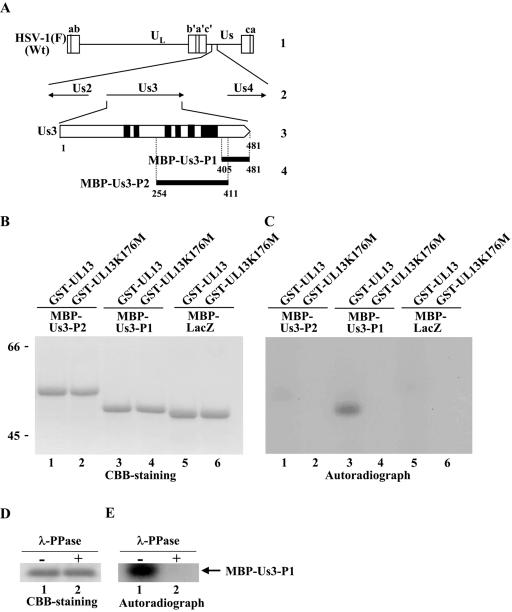
FIG. 2.
(A) Schematic diagram of the genome structures of wild-type (Wt) virus HSV-1(F) and the location of the Us3 gene. Line 1, linear representation of the HSV-1(F) genome. The unique sequences are represented as unique long (UL) and short (Us) domains, and the terminal repeats flanking them are shown as open rectangles with the designation above each repeat. Line 2, structure of the genome domain containing the Us2, Us3, and Us4 open reading frames. Line 3, structure of the Us3 open reading frame. The shaded areas represent subdomains I to VI, which are conserved in eukaryotic protein kinases (68). Line 4, the domains of the Us3 gene used in these studies to generate MBP-Us3 fusion proteins. (B) CBB-stained images of phosphorylated Us3. Purified MBP-Us3-P2 (lanes 1 and 2), MBP-Us3-P1 (lanes 3 and 4), and MBP-LacZ (lanes 5 and 6) incubated in kinase buffer containing [γ-32P]ATP and purified GST-UL13 (lanes 1, 3, and 5) or GST-UL13K176M (lanes 2, 4, and 6), separated on a denaturing gel, and stained with CBB. Molecular masses (kDa) are shown on the left. (C) Autoradiograph of the gel in panel B. (D) Purified MBP-Us3-P1 incubated in kinase buffer containing [γ-32P]ATP and purified GST-UL13 and then either mock treated (lane 1) or treated with λ-PPase (lane 2), separated on a denaturing gel, and stained with CBB. (E) Autoradiograph of the gel in panel D.