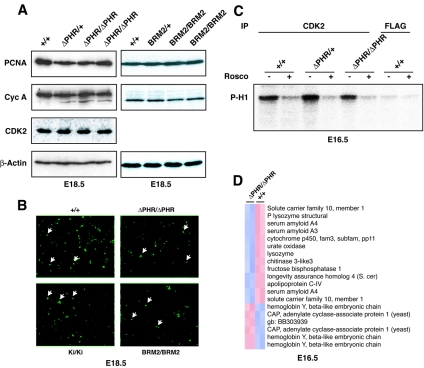
FIG. 6.
In vivo growth regulation during liver development in the ΔPHR homozygous mice. (A) Western blots of nuclear extracts isolated from fetal livers of +/+, ΔPHR/+, ΔPHR/ΔPHR, BRM2/+, and BRM2/BRM2 embryos were probed with antibodies raised against PCNA, cyclin A, CDK2, and β-actin. (B) BrdU incorporation in fetal liver (original magnification, 20×). A few labeled nuclei are indicated by arrows. The percentage of BrdU-labeled nuclei in the wild-type (+/+) mice, the ΔPHR homozygotes (ΔPHR/ΔPHR), the Wtki homozygotes (Ki/Ki), and the BRM2 homozygotes (BRM2/BRM2) ranged between 5 and 7% (approximately 600 nuclei were counted). (C) Phosphorylation of histone H1 (P-H1) was assayed using anti-CDK2- and anti-FLAG-precipitated liver lysates isolated from +/+, ΔPHR/+, or ΔPHR/ΔPHR embryos. The CDK2 inhibitor roscovitine (Rosco) was added in the lanes marked with a plus sign. (D) Transcriptional gene profiling of total fetal liver RNA derived from two ΔPHR/ΔPHR mice and two +/+ littermate controls. Hierarchical clustering of the 14 genes displays altered gene expression. Pearson correlation coefficients were >0.988 for the individual pairwise comparisons. S. cer, Saccharomyces cerevisiae; gb, GenBank.