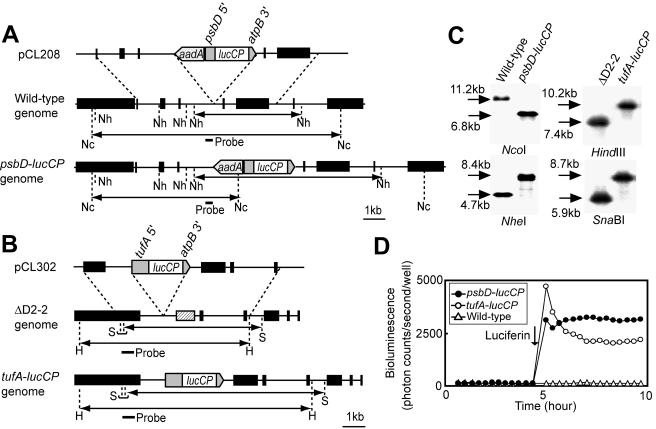
FIG. 2.
Construction of bioluminescence reporter strains. (A) Schematic representation of the reporter construct and the chloroplast genomes of psbD-lucCP and wild-type strains. psbD 5′ represents the promoter and 5′-UTR of the psbD gene; atpB 3′ represents the 3′-UTR of the atpB gene. The black boxes of the chloroplast genomes denote genes: from left to right, wendy, trnE2, psbH, psbN, psbT, psbB, trnD, and rpoA. The small bar indicates the location of the probe used for Southern blot analysis. The double-headed arrows indicate fragments expected to be detected by Southern blot analysis. Restriction sites: Nc, NcoI; Nh, NheI. (B) Schematic representation of the reporter construct and the chloroplast genomes of tufA-lucCP and ΔD2-2 strains. tufA 5′ represents the promoter and 5′-UTR of the tufA gene. The black boxes denote genes: from left to right, ORF2971, psbD (replaced with the 483-bp repeats [12] in the ΔD2-2 genome [hatched box]), psaA exon 2, psbJ, atpI, psaJ, and rps12. Restriction sites: H, HindIII; S, SnaBI. (C) Southern blot analysis of genomic DNAs. Genomic DNAs digested with the restriction enzyme were hybridized with the probes indicated in panels A and B. Sizes of detected bands are indicated. (D) Representative traces of bioluminescence from the wild-type and reporter strains. One hundred microliters of mid-log-phase cultures grown in TAP medium was transferred into individual wells of a 96-well microtiter plate, and bioluminescence was measured every 20 min with the automated bioluminescence-monitoring apparatus. The arrow indicates when luciferin was added (final concentration, 100 μM).