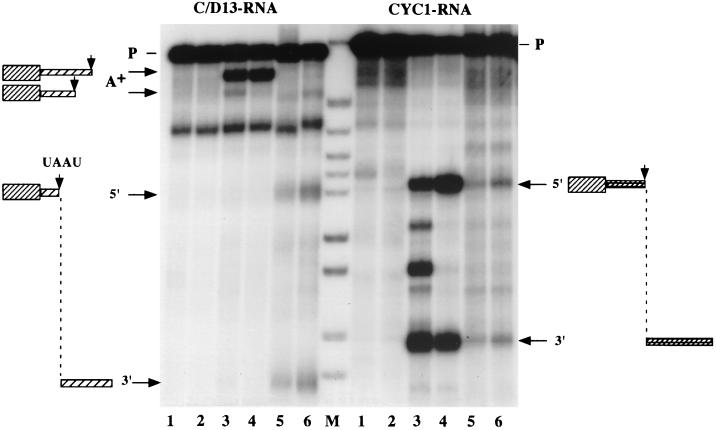
FIG. 8.
CPF, CF IA, and CF IB are not sufficient for correct cleavage of a box C/D snoRNA substrate at the in vivo processing site. The RNA precursors and processing products are indicated on the sides of the panels. Internally 32P-labeled RNAs were incubated with CF IA (lanes 2), CF IA and CPF (lane 3), CF IA, CPF, and 100 ng of GST-Nab4p (lanes 4), processing extract (lanes 5), or whole-cell extract (lanes 6). Input RNA alone was loaded in lanes 1. RNAs were resolved on a 6% polyacrylamide-8.3 M urea gel. HpaII-digested pBR322 fragments were 5"-end labeled and served as markers (lane M). A+ bands identify the sites that become polyadenylated, while the 5" and 3" bands are the 5" and 3" cutoff products when cleavage occurs at the UA"AU sequence, which is utilized as the entry site for 3" processing in vivo.