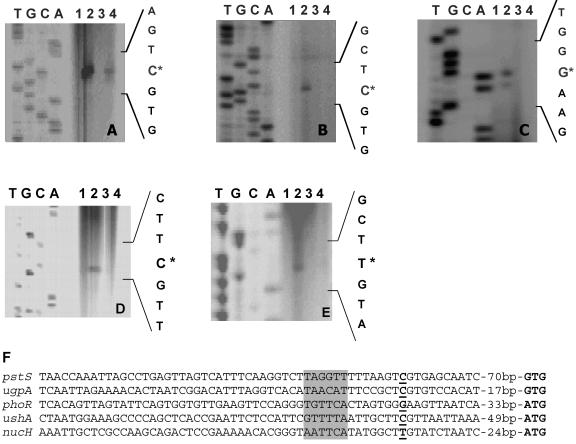
FIG. 4.
Comparison of mRNA levels and determination of the transcriptional start sites of the C. glutamicum genes pstS (A), ugpA (B), phoR (C), ushA (D), and nucH (E) by primer extension analysis. The reverse transcriptase reactions were performed with the oligonucleotides pstS_prext2, ugpA_prext2, phoR_prext1b, ushA80prext, and nucH90prext for these four genes, respectively, and 20 μg of total RNA was isolated from the following strains: the wild type grown under phosphate excess (lane 1); the wild type 60 min (A and B), 10 min (C), or 90 min (D and E) after a shift from 13 mM Pi to 0.065 mM Pi (lane 2); the ΔphoRS mutant grown under Pi excess (lane 3); and the ΔphoRS mutant 60 min (A and B), 10 min (C), or 90 min (D and E) after a shift from 13 mM Pi to 0.065 mM Pi (lane 4). The transcriptional start sites are indicated by asterisks. The corresponding sequencing reactions were generated by using the same IRD-800-labeled oligonucleotide as in the primer extension reactions as well as PCR products which cover the region of the respective transcriptional start site as the template DNA. In panel F, the promoter regions derived from the primer extension studies were aligned. The transcriptional start points are shown in bold and underlined, the proposed start codons are indicated in bold, and putative −10 regions are shaded in gray.