Abstract
Regions with tandemly arranged leucine-rich repeats (LRRs) have been found in many prokaryotic and eukaryotic proteins, in which they provide a remarkably versatile framework for the formation of ligand-binding sites. Bacterial LRR proteins include the recently described Slr protein of Streptococcus pyogenes, which is related to internalin A of Listeria monocytogenes. Here, we show that strains of the human pathogen Streptococcus agalactiae express a protein, designated Blr, which together with Slr defines a family of internalin A-related streptococcal LRR proteins. Analysis with specific antibodies demonstrated that Blr is largely inaccessible on S. agalactiae grown in vitro, but surface exposure was increased ∼100-fold on mutants lacking polysaccharide capsule. In S. pyogenes, surface exposure of Slr was not affected in a mutant lacking hyaluronic acid capsule but was increased >20-fold in mutants lacking M protein or protein F. Thus, both Blr and Slr are efficiently camouflaged by other surface structures on bacteria grown in vitro. When Blr and Slr exposed on the bacterial surface were compared, they exhibited only little immunological cross-reactivity, in spite of extensive residue identity, suggesting that their surface-exposed parts have been under evolutionary pressure to diverge functionally and/or antigenically. These data identify a family of immunologically diverse streptococcal LRR proteins that show unexpected complexity in their interactions with other bacterial surface components.
Regions with tandemly arranged leucine-rich repeats (LRRs) occur in many proteins and have been implicated in interactions with different ligands (25). The wedge-shaped LRRs in these regions give rise to a curved structure, which provides a versatile framework for the formation of a ligand-binding surface on the concave side. Each individual LRR is 20 to 29 amino acids long and includes the 11-residue consensus sequence LXXLXLXXNXL (where X is any amino acid). The leucines can be replaced by other amino acids with hydrophobic side chains, most frequently isoleucine or valine but also phenylalanine, whereas the asparagine is sometimes replaced with a cysteine or a threonine. Among human LRR-containing proteins are the TLR (Toll-like receptor) proteins, the NOD (nucleotide-binding oligomerization domain) proteins, and other proteins implicated in the detection of pathogen-derived molecules and generation of an inflammatory response (11, 19, 30).
Several pathogenic bacteria, both gram positive and gram negative, express virulence factors with LRR regions. These bacterial proteins include internalins A and B (InlA and InlB) of Listeria monocytogenes (6), the YopM protein of Yersinia pestis (8), the IpaH family of proteins in Shigella flexneri (60), and the SspH proteins of Salmonella enterica serovar Typhimurium (33). The classical example of a bacterial LRR protein is the L. monocytogenes protein InlA, which binds to human E-cadherin and promotes adhesion to and invasion of cells in the intestinal epithelium (10, 27, 32).
Recent work has identified an InlA-related LRR protein, designated Slr (Spy1361), in the gram-positive bacterium Streptococcus pyogenes (group A streptococcus), the cause of acute pharyngitis (“strep throat”), skin infections, streptococcal toxic shock syndrome, and several other diseases (45, 46). This surface-localized protein, which has an LRR region in the C-terminal part, has the characteristics of a lipoprotein, implying that it is attached to the bacterial cell membrane via a cysteine residue in the N-terminal region. In contrast, InlA is covalently attached to the cell wall via an LPXTG sequence in the C-terminal part (4), while the LRR region is located in the N-terminal half, implying that the LRR region is located most distally from the cell wall in both of these proteins (Fig. 1A).
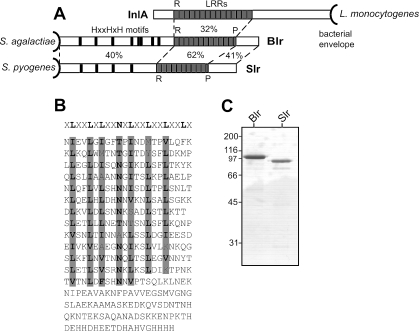
FIG. 1.
Analysis of the Blr protein of S. agalactiae. (A) The Blr protein exhibits extensive residue identity to Slr of S. pyogenes (46), and both of these predicted lipoproteins are related to L. monocytogenes InlA, which is attached to the wall via an LPXTG motif. Amino acid residue identities of different regions are indicated in percents between the proteins. In all three proteins, the regions with LRRs are probably located distally to the bacterial surface. There are 12.5 LRRs in Blr, 10.5 LRRs in Slr, and 15 LRRs in InlA. The positions of histidine triad motifs (HXXHXH) in the N-terminal part of Blr and Slr are indicated. R, repeat; P, partial repeat. (B) Amino acid sequence of the C-terminal part of the Blr protein, including the LRR domain. The consensus sequence of LRRs in L. monocytogenes InlA is indicated in bold at the top, and the corresponding residues in the LRRs of Blr are highlighted in gray. In Blr, each LRR has a length of 22 residues, with the exception of the seventh repeat, which is one residue shorter. (C) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of purified recombinant Blr and Slr. Molecular weight markers (in thousands) are noted at the left of the gel.
Our work was focused on the human pathogen Streptococcus agalactiae (group B streptococcus), the most important cause of life-threatening diseases like pneumonia, sepsis, and meningitis in newborns (7). Although the two streptococcal species S. agalactiae and S. pyogenes cause different diseases and express many different virulence factors, some surface proteins expressed by these pathogens are closely related (29). This situation prompted us to analyze whether S. agalactiae expresses an LRR protein related to Slr. Here, we describe such a protein, designated Blr (for group B, leucine rich), and compare it with the Slr protein of S. pyogenes. These studies demonstrated that Blr and Slr define a family of streptococcal LRR proteins that exhibit extensive amino acid residue identity but nevertheless show only weak immunological cross-reactivity when exposed on the bacterial surface. Remarkably, access of antibodies to Blr or Slr was strongly reduced by other surface components on bacteria grown under laboratory conditions. Access to Blr was inhibited by the polysaccharide capsule of S. agalactiae, and access to Slr was inhibited by M protein and protein F of S. pyogenes. These interactions are likely to affect the in vivo function of the streptococcal proteins and are of general interest for studies of bacterial surface proteins.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
S. agalactiae BM110 is a capsular serotype III strain of a putative high-virulence clone (29, 35, 52). The S. agalactiae type III strain COH1 and its isogenic acapsular mutant, COH1-13 (47), were from C. Rubens (Children's Hospital, Seattle, Wash.), and the S. agalactiae type II strain 1954/92 was from R. Facklam (Centers for Disease Control and Prevention, Atlanta, Ga.). Strains of S. agalactiae representing the nine known capsular serotypes were available in our laboratory. The S. pyogenes M6 wild-type strain JRS4 and its M-negative derivative, JRS145, were from J. R. Scott (Emory University, Atlanta, Ga.) (5). The protein-F-negative JRS4 mutant SAM1 and the double mutant SAM2, which lacks both M6 and protein F, were from E. Hanski (Hebrew University, Jerusalem, Israel) (16). The S. pyogenes M5 Manfredo strain (34) and its M-negative mutant, ΔM5, have been described previously (20). The encapsulated S. pyogenes M18 strain 87-282 and its capsule-negative mutant, TX72, were from M. R. Wessels (Children's Hospital, Boston, Mass.) (63). All S. agalactiae strains were grown in Todd-Hewitt broth (TH) at 37°C without shaking; all S. pyogenes strains were grown in TH supplemented with 0.2% yeast extract (THY) in 5% CO2 at 37°C without shaking. For construction of bacterial mutants, we employed plasmid pJRS233, which carries an erythromycin (Ery) resistance gene and exhibits temperature-sensitive replication in gram-positive bacteria, allowing selection through homologous recombination (42). Plasmid pFW14 (provided by Ulrika Ringdahl, Lund University) is a derivative of pFW8 (43) in which cat has been fused with the promoter region and termination transcription signals from the spectinomycin resistance gene aad9, resulting in constitutive expression of cat.
Construction of a Blr-negative S. agalactiae mutant.
A Blr-negative mutant, designated Δblr-36, was constructed from strain BM110. A derivative of pJRS233 was used to replace the blr gene in strain BM110 with the kanamycin resistance cassette ΩKm2 (41). The pJRS233 derivative, designated pJWΔblr, carried the ΩKm2 cassette surrounded by chromosomal regions upstream and downstream of the blr gene, allowing replacement of the blr gene by homologous recombination. For construction of pJWΔblr, a 1,020-bp region immediately downstream of blr was first amplified by PCR, the PCR product was digested with BamHI and EcoRI (for which recognition sequences had been introduced through the primers), and the fragment was ligated into BamHI/EcoRI-cleaved pBR322. Similarly, the 1,014-bp region immediately upstream of blr was amplified by PCR and the resulting PCR fragment was digested with SalI and BamHI, for which recognition sequences had been introduced through the primers. This fragment was ligated into the pBR derivative carrying the downstream sequence (cleaved with SalI and BamHI), generating a plasmid in which the regions upstream and downstream of blr are separated by a BamHI site. This plasmid was cleaved with BamHI and ligated to BamHI-cleaved ΩKm2, generating plasmid pBRΔblr. Finally, the whole insert in pBRΔblr was amplified by PCR, cleaved with SalI and XbaI (recognition sequences introduced through the primers), and ligated into SalI/XbaI-cleaved pJRS233, generating plasmid pJWΔblr. This construct was transformed into strain BM110, and the mutant (BM110 Δblr-36) was recovered after homologous recombination (42). The structure of the mutant was verified by PCR. Analysis by reverse transcriptase PCR demonstrated that the Δblr mutation did not have a polar effect on transcription of the gene located downstream (data not shown).
Construction of a Rib-negative S. agalactiae mutant.
A Rib-negative mutant, designated Rm69, was derived from strain BM110. An insert harboring the rib gene and the flanking chromosomal regions upstream (∼2.2 kb) and downstream (∼2.0 kb) was recovered from a pUC19 derivative (61; unpublished data) by cleavage with SalI and HindIII, and the insert was ligated into SalI/HindIII-cleaved pJRS233, generating plasmid pTArib. Cleavage of plasmid pTArib with BglII could be used to remove a large part of the central repeat region in rib because each repeat unit contains a BglII cleavage site (61). BamHI-cleaved ΩKm2 was therefore ligated into BglII-cleaved pTArib, generating plasmid pTA110, in which most of the central repeat region in rib has been replaced with ΩKm2. This construct was transformed into strain BM110, and a Rib-negative mutant was recovered after homologous recombination (42). The structure of the mutant was verified by PCR.
Construction of capsule-negative S. agalactiae mutants.
Capsule-negative S. agalactiae mutants were derived from the wild-type strain BM110 and its Rib-negative mutant, Rm69. The construction of the mutants employed a pJRS233 derivative, in which the chloramphenicol (Cm) resistance gene cat (43) was surrounded by regions derived from the cps operon of S. agalactiae serotype III (47). For construction of this plasmid, a region of the cps operon (1,308 bp), including 23 bp of the 3′ end of cpsC, an intergenic region of 471 bp, and most of cpsD (814 bp), was amplified by PCR. The resulting PCR product was cleaved with SalI and XbaI (recognition sequences introduced through the primers) and cloned into SalI/Xba-cleaved pJRS233. This plasmid was designated pTAcpsD. The cat gene was amplified by PCR from plasmid pFW14 and ligated into a naturally occurring HindIII site in the cpsD gene, generating plasmid pTAcpsDcat. In this plasmid, the cat gene is flanked by S. agalactiae regions 745 bp upstream and 563 bp downstream of the HindIII site. Plasmid pTAcpsDcat was transformed into strains BM110 and Rm69, and mutants in which the insert had been introduced by homologous recombination were sought. For this purpose, transformants were inoculated into THY containing erythromycin (1 μg/ml), grown overnight at 30°C, and subsequently grown twice in THY containing chloramphenicol (5 μg/ml) at 37°C. Colonies with a Cmr Erys phenotype were isolated and characterized. Capsule-negative mutants of both BM110 and Rm69 were obtained and were designated BM110-22 and Rm69-16, respectively. When analyzed by PCR, the capsule-negative mutant Rm69-16 was shown to represent a double crossover, as expected. In contrast, the capsule-negative mutant BM110-22 was unexpectedly found to be a single-crossover mutant, although it was Erys. However, because this mutant had the correct phenotype, it could be used for the experiments reported here. Repeated attempts to isolate a double-crossover, capsule-negative mutant from BM110 were unsuccessful.
Purification of recombinant Blr and Slr.
The blr gene, except the first 69 nucleotides that encode the signal sequence and the putative lipobox, was amplified by PCR from S. agalactiae strain BM110. The product was digested with BamHI and EcoRI, using recognition sequences introduced through the primers, and ligated into BamHI/EcoRI-cleaved plasmid pGEX-6P-2 (Amersham), yielding the expression vector pGEX-blr. The slr gene, except the first 69 nucleotides, was similarly amplified from S. pyogenes strain M5 Manfredo, for which the genome has been sequenced (http://www.sanger.ac.uk/Projects/S_pyogenes/), and the amplified slr gene was inserted into pGEX-6P-2, yielding the expression vector pGEX-slr. Overnight LB cultures of Escherichia coli BL21 harboring either of the two expression vectors were diluted 200-fold in fresh LB containing ampicillin (100 μg/ml) and cultured at 37°C with rotation (200 rpm) to an A600 of ∼0.5. The culture was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) (final concentration of 1 mM), and incubation was continued for 2 h. The bacteria were collected by centrifugation, washed and suspended in phosphate-buffered saline (PBS), and lysed by repeated freezing and thawing. After removal of cellular debris by centrifugation, the preparation was partially purified by DEAE chromatography before it was applied to a 5-ml GSTrap column (Amersham). The purification and the removal of the glutathione S-transferase tag were performed according to the manufacturer's instructions, except that the binding buffer was 50 mM Tris-HCl, pH 7.0. Fractions containing purified protein were pooled and dialyzed against PBS.
Antisera.
Antisera against purified Blr and Slr were raised in rabbits as described previously (2). About 70 μg of protein in complete Freund’s adjuvant was used for the first immunization, and boosters of about 35 μg were given at 2- to 3-week intervals, using incomplete Freund’s adjuvant. Antiserum to S. agalactiae type III capsule, conjugated to tetanus toxoid, was the kind gift of D. L. Kasper (Boston, Mass.).
Binding assays with antisera and bacteria.
The bacteria in cultures incubated overnight (∼16 h) were washed twice in PBS with 0.02% NaN3 and 0.05% Tween 20 (PBSAT) and resuspended in PBSAT to 1 × 109 CFU/ml. Samples (200 μl) were distributed to a series of tubes, to which antiserum (diluted in PBSAT as indicated) was added. After incubation at room temperature for 1 h with gentle shaking, the samples were centrifuged and the bacteria were washed twice with PBSAT. For detection of bound antibodies, 125I-labeled staphylococcal protein A (∼10,000 cpm in 200 μl PBSAT) was added to each bacterial pellet and the incubation was continued for 30 min with gentle shaking. Following washing with PBSAT, bound protein A was measured in a γ-counter. Binding is presented as percent radiolabeled protein A added. Because a standard amount of protein A was added to each bacterial sample to detect bound antibodies, binding of protein A may reach a maximal level at lower antiserum dilutions and decreases at higher dilutions. Some of the protein A will have lost binding ability during the radiolabeling, explaining why maximal binding is always <100%. The antibody binding was not caused by Fc-binding bacterial surface proteins, because the streptococcal strains used here do not express such proteins. Moreover, preimmune serum did not react with the strains, as indicated in the figure legends.
Immunofluorescence and phase-contrast microscopy.
For detection of surface-exposed Blr by immunofluorescence, a 10-ml culture of the S. agalactiae strain indicated was grown to late logarithmic phase (A620 of ∼0.8). The bacteria were washed twice with PBSAT and resuspended to 2 × 109 CFU/ml in the same buffer. Samples (100 μl) were distributed to a series of tubes, to which 10 μl rabbit anti-Blr serum (diluted in PBSAT, final dilution of 1/100) was added. Control samples were incubated with preimmune serum. After incubation at room temperature for 1 h with gentle shaking, the samples were centrifuged and the bacteria were washed twice with PBSAT. To detect bound antibodies, Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin G (Molecular Probes) was added and the incubation was continued for 20 min with gentle shaking. After washes with PBSAT, the samples were analyzed by phase-contrast and immunofluorescence microscopy. Fading of the fluorochrome was reduced by the use of a SlowFade light antifade kit (Molecular Probes).
Analysis of cross-reactivity between Blr and Slr.
The cross-reactivity between pure Slr and Blr was analyzed by enzyme-linked immunosorbent assay (ELISA). The wells of microtiter plates (Maxisorp; Nunc) were coated with protein by overnight incubation with 50 μl carbonate buffer, pH 9.5, containing Blr or Slr (1 μg/ml). The plates were washed three times with PBSAT and blocked by incubation for 1 h with PBSAT containing 1% bovine serum albumin. After three washes with PBSAT, 50 μl rabbit antiserum, diluted in blocking buffer as indicated, was added to each well. The plates were incubated for 2 h and washed three times with PBSAT. To detect antibody binding, 50 μl protein A conjugated to biotin (0.33 μg/ml in blocking buffer) was added to each well and the incubation was continued for 1 h. Following washing, 50 μl streptavidin conjugated to horseradish peroxidase (HRP) (0.10 μg/ml in blocking buffer) was added to each well. After 30 min of incubation and further washing, 50 μl of substrate solution (o-phenylenediamine dihydrochloride, Fast P9187; Sigma) was added to each well. The signal was detected with an ELISA plate reader at A450. All incubations were performed at room temperature, except the coating step, which was performed at 4°C. The cross-reactivity of Blr and Slr present on the bacterial surface was analyzed by performing binding assays with antisera and washed bacteria, as described above.
Analysis of bacterial extracts by Western blotting.
Cultures of S. agalactiae and S. pyogenes incubated overnight (∼16 h) were washed twice in 50 mM Tris-HCl, pH 7.3, and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer to 2 × 1010 CFU/ml. After the preparation was boiled for 10 min and debris was removed by centrifugation, the extracts were analyzed by Western blotting, using rabbit anti-Blr or anti-Slr as the probe. Bound antibodies were detected with protein A-conjugated biotin followed by streptavidin conjugated to HRP, and HRP was detected by incubation with SuperSignal West Pico substrate (Pierce) for 5 min.
Mouse infection model for analysis of virulence.
Log-phase cultures of strain BM110 and its nonpolar Blr-negative mutant, Δblr-36, were diluted in TH, and 0.5 ml (3 × 105 CFU) was used for intraperitoneal (i.p.) infection of C3H/HeN mice. For the wild-type strain, preliminary analysis had indicated that the number of CFU used corresponded to a 90% lethal dose. Deaths were recorded regularly for 96 h. The experiments were approved by the Lund-Malmö review board on animal studies.
Analysis of the immune response in infected mice.
C3H/HeN mice were injected i.p. with a sublethal dose (∼1 × 104 CFU) of live S. agalactiae bacteria of the strain indicated. The mice were sacrificed after 1 month, and sera were collected and analyzed for the presence of anti-Blr antibodies. For this purpose, the wells of microtiter plates (Falcon 9612; Becton Dickinson) were coated overnight with Blr (1 μg/ml in PBS), washed with PBSAT, and blocked with the same buffer for 1 h. Following washing, mouse serum (50 μl, diluted as indicated in PBSAT) was added to each well, and the plates were incubated for 2 h. After the wells were washed with PBSAT, 50 μl rabbit anti-mouse immunoglobulin (Z0259, diluted 2,000-fold in PBSAT; DakoCytomation) was added to each well and the incubation was continued for 1 h. After further washing, bound rabbit antibodies were detected with 125I-labeled protein G (∼10,000 cpm in 50 μl PBSAT). All incubations were performed at room temperature with gentle shaking, except the coating step, which was performed at 4°C without shaking.
Nucleotide sequence accession number.
The GenBank accession number for the nucleotide sequence of the blr gene of strain BM110 is DQ242614.
RESULTS
Sequence comparison of Blr and Slr and purification of recombinant proteins.
Through searches of genomic databases, we found that a homolog of the S. pyogenes slr gene was present in the two published genomes of S. agalactiae (12, 58), as also noted by Sutcliffe and Harrington (54). Preliminary work showed that this homolog, designated blr, was also present in S. agalactiae strain BM110, which was used for most of the work described here because it is a member of a putative high-virulence clone of the clinically important serotype III (29, 35). Sequencing of blr in BM110 and comparison with the blr genes in the two published S. agalactiae genomes demonstrated that the three genes encode predicted Blr proteins with 98% amino acid residue identity. In each strain, the blr gene encodes an 877-residue polypeptide (including signal sequence), with a predicted molecular weight of 97,590 in strain BM110 (Fig. 1A). The N-terminal part of Blr contains a predicted lipobox (residues 19 to 23), which is required for covalent attachment of a lipoprotein to the cell membrane (54, 55), while the C-terminal part, which includes 12.5 LRRs, is predicted to be located distally to the surface. Each LRR contains 22 amino acid residues, with the exception of the seventh repeat, which is one residue shorter.
The N-terminal parts of Blr and Slr show 40% residue identity, and the LRR regions show 62% identity (Fig. 1A). While Blr contains 12.5 LRRs, there are 10.5 LRRs in the Slr protein (46). As expected from the work with Slr (46), Blr shows sequence similarity to InlA, with 32% residue identity between the LRR regions. Because InlA is attached to the bacterial cell wall via a C-terminal LPXTG motif (4), the LRR region is predicted to be located distally to the bacterial surface in all three proteins, a location that may favor interactions with ligands. Interestingly, the LRRs of InlA, Blr, and Slr have the same length, 22 residues, except one LRR with a length of 21 residues. In InlA, it is the sixth repeat that only contains 21 residues (49). The N-terminal part of Blr includes eight histidine triad motifs (HXXHXH), while four such motifs were found in Slr (Fig. 1A). The same motif, for which no function has been identified, has been described for several surface proteins of Streptococcus pneumoniae (1, 64). Together, the similarities between Blr and Slr demonstrate that these two proteins are members of a family of streptococcal proteins with LRRs.
The gene encoding Slr was present in all strains of S. pyogenes analyzed by Reid et al. (46). To analyze different S. agalactiae strains for the presence of the blr gene, genomic DNA from 17 isolates, including at least one strain for each of the nine known capsular serotypes, was analyzed by PCR. The blr gene was present in all 17 isolates tested (data not shown), implying that most, if not all, strains of S. agalactiae harbor the blr gene.
Interestingly, Blr and Slr show homology not only to internalin A of L. monocytogenes but also to a putative internalin-related surface protein in Bacillus anthracis (44). In contrast, searches of databases did not reveal any predicted LRR proteins closely related to Blr or Slr in the gram-positive cocci Staphylococcus aureus, Streptococcus pneumoniae, and Enterococcus faecalis.
For purification of the Blr and Slr proteins, DNA fragments of blr and slr corresponding to amino acids 24 to 877 and 24 to 792, respectively, were cloned into the expression vector pGEX-6P-2. In these DNA fragments, the first 69 nucleotides, corresponding to the first 23 amino acids, were deliberately omitted to avoid potential toxicity due to the signal sequence and the lipobox (13). Each protein was overexpressed in E. coli BL21 and purified to apparent homogeneity (Fig. 1C), and antisera were raised in rabbits.
The Blr protein is located on the S. agalactiae surface but is camouflaged by the capsule.
The presence of a putative lipobox in Blr and the report that the related Slr protein is exposed on the surface of S. pyogenes (46) suggested that Blr would be exposed on the surface of S. agalactiae. However, antibodies to Blr reacted only weakly with washed whole S. agalactiae bacteria (see below). This result was not due to low titer of the antiserum or release of the protein into the culture supernatant (data not shown).
We hypothesized that the weak reactivity of S. agalactiae bacteria with anti-Blr was not due to low-level expression of the protein but to camouflaging by other surface components. The capsule was of particular interest in this context, because it has been suggested that it may reduce the exposure of S. agalactiae surface proteins (57). Surface exposure of Blr in a capsule-negative BM110 mutant was therefore analyzed. Because BM110 and most other S. agalactiae serotype III strains express the major surface protein Rib (29, 52, 61), we also analyzed whether lack of this protein affected the ability of antibodies to detect Blr. Moreover, surface exposure of Blr was analyzed in a double mutant lacking both capsule and Rib.
The BM110 mutants were constructed as described in Materials and Methods. Characterization of the mutants with specific antibodies demonstrated that they had the expected properties (Fig. 2). Thus, the capsule-negative mutant (BM110-22) reacted with anti-Rib but not with anticapsule antibodies (Fig. 2B), the Rib-negative mutant (Rm69) reacted only with anticapsule antibodies (Fig. 2C), and the double mutant (Rm69-16) lacked reactivity with both antisera (Fig. 2D).
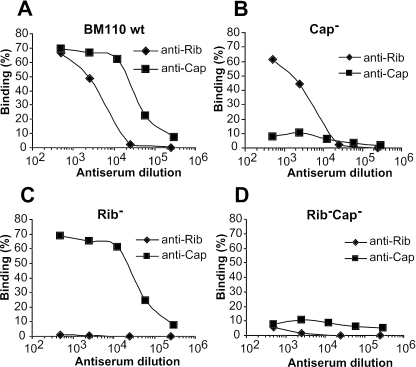
FIG. 2.
Characterization of mutants of S. agalactiae strain BM110 that lack polysaccharide capsule and/or protein Rib. (A) Binding of anticapsular and anti-Rib antibodies to the BM110 wild type (wt). A series of identical bacterial samples were mixed with antisera and diluted as indicated, and bound antibodies were detected by the addition of a standard amount of radiolabeled protein A, as described in Materials and Methods. Binding is presented as percentage of protein A added, explaining why binding may reach a maximal level at lower antibody dilutions and decreases at higher dilutions. (B, C, and D) Binding of anticapsular and anti-Rib antibodies to the BM110 mutants indicated. The capsule-negative mutant (Cap−) is strain BM110-22, the Rib-negative mutant is strain Rm69, and the double mutant is strain Rm69-16. All experiments were performed three times with triplicate samples, and each panel shows representative data from one experiment. Binding observed with preimmune serum (<8%) has been subtracted.
Analysis of surface exposure of Blr on the capsule- and Rib-negative mutants yielded striking results (Fig. 3A). Anti-Blr antibodies reacted well with the capsule-negative mutant, but to obtain similar reactivity with the wild-type strain, ∼100-fold more antibodies were required, indicating that ∼100-fold less Blr was exposed on the surface of the latter strain. Compared to the capsule-negative mutant, an additional ∼4-fold increase in Blr exposure was observed for the double mutant lacking both capsule and Rib. However, exposure of Blr was not increased with the mutant lacking only Rib. The strongly increased reactivity of anti-Blr with the two strains lacking capsule was not due to exposure of a cross-reacting protein, because the anti-Blr serum is highly specific and detects only Blr in extracts of S. agalactiae (see below). These data indicate that the capsule camouflages surface-localized Blr and also suggest that Rib contributes to the masking, at least in the absence of capsule. This result was not unique to strain BM110, because strongly increased reactivity with anti-Blr was also observed with a capsule-negative mutant of another S. agalactiae strain, COH1 (47) (Fig. 3B).
FIG. 3.
The capsule of S. agalactiae inhibits binding of antibodies to Blr but not to protein Rib. (A) Binding of anti-Blr antibodies to S. agalactiae strain BM110 wild type (wt) and mutants of this strain. For each bacterial strain, a series of identical samples were mixed with antisera and diluted as indicated, and bound antibodies were detected as described in Materials and Methods. The mutants used were those described in the legend to Fig. 2. (B) Binding of anti-Blr antibodies to S. agalactiae strain COH1 and its acapsular mutant, COH1-13 (Cap−). (C) Binding of anti-Blr antibodies to S. agalactiae wt strain BM110 and its acapsular mutant, BM110-22 (Cap−), analyzed by immunofluorescence. A control with preimmune (preimm) serum is shown for BM110-22. Bound antibodies were detected as described in Materials and Methods, and representative images from one experiment are presented. For each analysis, the same field is shown in the phase-contrast panel (Phase) and in the immunofluorescence panel (IF). (D) Western blot analysis of bacterial extracts, using anti-Blr as the probe. The extracts were prepared from S. agalactiae BM110 wild type, its Rib- and capsule-negative double mutant (Rm69-16), and the Blr-negative BM110 mutant, Δblr-36. Molecular weight markers (in thousands) are noted at the left of blots. (E) Lack of capsule does not affect surface exposure of protein Rib. Anti-Rib antibodies, diluted as indicated, were incubated with BM110 wild type and its acapsular mutant, BM110-22 (Cap−). This panel was derived from data presented in Fig. 2A and B. The binding experiments shown in panels A, B, and E were performed three times with triplicate samples, and each panel represents data from one experiment. Binding observed with preimmune serum (<8%) has been subtracted.
To verify the results obtained in binding assays, in which radiolabeled protein A was used to detect bound antibodies, the surface exposure of Blr was also analyzed by immunofluorescence (Fig. 3C). The wild-type strain BM110 showed no reactivity with anti-Blr antibodies, while the acapsular mutant, BM110-22, reacted strongly with these antibodies. This reactivity was not unspecific, because the acapsular mutant did not react with preimmune serum. These data confirm that Blr is a surface protein, which is poorly accessible in the presence of the capsule.
The increased surface exposure of Blr with the capsule-negative mutants could be explained by camouflaging, but it could not be excluded that it reflected upregulation of Blr synthesis through an unknown mechanism. To analyze this possibility, we compared the total amount of Blr protein present in extracts of the wild-type strain BM110 and in the double mutant lacking capsule and Rib. Analysis of the extracts by Western blotting with anti-Blr as the probe indicated that the two extracts contained similar amounts of Blr (Fig. 3D). The protein detected in this blot was Blr and not a cross-reacting protein, because no signal was obtained for an extract prepared from a Blr-negative bacterial mutant (Fig. 3D, third lane). (See Materials and Methods for the construction of the Blr-negative mutant). These data indicate that the surface-localized Blr protein is indeed camouflaged by other cell wall components and in particular by the capsule.
To analyze whether the capsule causes camouflaging of surface proteins different from Blr, we used anti-Rib to compare the surface accessibilities of Rib in strain BM110 and its capsule-negative mutant (Fig. 3E). In contrast to Blr, exposure of Rib was not affected by the absence of capsule. This result does not exclude that the capsule camouflages other proteins but shows that it selectively camouflages Blr compared to Rib. Because Blr and Rib were detected by identical techniques, using rabbit antibodies, these data also show that the effect of the capsule on the binding of anti-Blr was specific.
The Slr protein of S. pyogenes is also camouflaged: roles of M protein and protein F.
The camouflaging of Blr in S. agalactiae suggested that a similar situation might prevail for Slr in S. pyogenes. Indeed, preliminary tests indicated that anti-Slr antibodies reacted only weakly with S. pyogenes wild-type bacteria. Because of the dramatic effect of the capsule in S. agalactiae, it seemed possible that the hyaluronic acid capsule that is expressed by at least some strains of S. pyogenes (62) might affect surface exposure of Slr. This hypothesis was analyzed with a capsule-producing (mucoid) strain of the clinically important serotype M18 and a capsule-negative mutant of that strain (63). Interestingly, absence of the hyaluronic acid capsule had no effect on surface exposure of Slr, as judged by the ability of antibodies to detect Slr (data not shown). This result suggests that the capsules of S. agalactiae and S. pyogenes differ in ability to mask the Blr and Slr proteins, respectively. Of note, the capsules of these two bacterial species are biochemically unrelated.
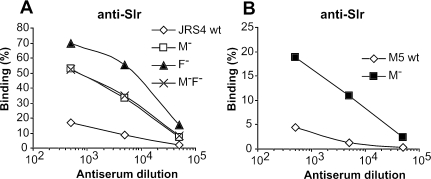
Although the hyaluronic acid capsule did not affect surface exposure of Slr, it seemed possible that Slr might be camouflaged by other surface components. Analysis of this problem was focused on two S. pyogenes surface proteins, the antiphagocytic M protein and the fibronectin-binding protein F (also known as Sfb1) (9, 16, 56). These two proteins were chosen for study because they are encoded in complex loci that play important parts in virulence, the emm and FCT loci, respectively, and because M protein, in particular, is a major component of the S. pyogenes cell envelope (3, 9, 36). The roles of M protein and protein F for surface exposure of Slr were studied with the M6 system, in which single mutants, lacking either protein, and a double mutant were available (5, 16). Surface exposure of Slr was increased more than 20-fold on all three strains and was even higher on the protein-F-negative mutant than on the other two mutant strains (Fig. 4A). Thus, absence of both proteins did not have a cumulative effect on the surface exposure of Slr. The detection of increased amounts of Slr on the mutants was not due to increased synthesis of the protein, because extracts of wild-type bacteria and of the mutants contained similar amounts of Slr, as demonstrated by Western blot analysis (data not shown). These results indicate that both M protein and protein F are required for camouflaging of Slr in the M6 strain of S. pyogenes. This result was not unique to the M6 system, because strongly increased exposure of Slr was also observed for an M-negative mutant derived from an M5 strain (Fig. 4B).
FIG. 4.
Two surface proteins of S. pyogenes, M protein and protein F, inhibit binding of antibodies to Slr. (A) Mutants of S. pyogenes strain JRS4 (of serotype M6) were analyzed for ability to bind anti-Slr. For each strain, a series of identical samples were mixed with antisera and diluted as indicated, and bound antibodies were detected as described in Materials and Methods. M− refers to the M6-negative strain JRS145, F− refers to the protein F-negative strain SAM1, and M−F− refers to the double mutant SAM2. (B) Binding of anti-Slr to S. pyogenes strain M5 Manfredo and its M-negative mutant, ΔM5 (M−). Each experiment was performed three times with triplicate samples, and representative data from one experiment are shown. Binding observed with preimmune serum (<9%) has been subtracted. wt, wild type.
Immunological cross-reactivity between Blr and Slr.
Because Blr and Slr exhibit extensive residue identity, it was of interest to analyze possible immunological cross-reactivity. It was not obvious that these two proteins would cross-react, as shown by analysis of the two S. agalactiae surface proteins Rib and α, which exhibit ∼50% residue identity but show little or no immunological cross-reactivity (52, 61).
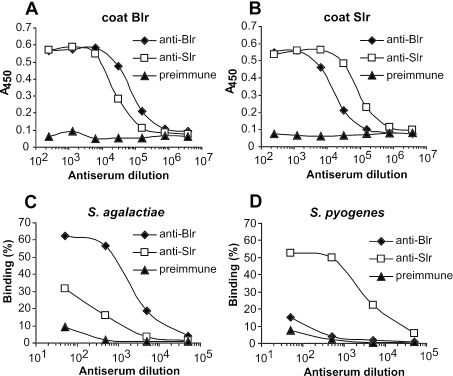
For examination of the immunological relationship between pure Blr and Slr, the proteins were immobilized in microtiter wells and analyzed for reactivity with anti-Blr and anti-Slr, using antisera that had the same titer against the homologous antigen (Fig. 5A and B). Both antisera reacted well not only with the homologous antigen but also with the heterologous protein, although the titer was highest against the homologous antigen. The greatest difference was observed for anti-Blr, but the reactivity of this antiserum was only ∼4-fold lower against the heterologous antigen. Thus, the two proteins showed strong cross-reactivity under these conditions. To analyze whether similar cross-reactivity would be observed under more physiological conditions, the two antisera were analyzed for reactivity with Blr and Slr expressed on the surface of bacteria. Because the proteins are largely camouflaged by other surface components on the wild-type strains, this analysis was performed with the S. agalactiae double mutant lacking capsule and Rib and with the S. pyogenes double mutant lacking M protein and protein F. Surprisingly, the cross-reactivity was very limited in this case (Fig. 5C and D). Although anti-Blr and anti-Slr had similar titers against bacteria expressing the homologous antigen, the titer against bacteria expressing the heterologous antigen was reduced >20-fold for anti-Slr and >100-fold for anti-Blr. Thus, most of the cross-reacting epitopes that were detected on pure proteins were not accessible on the proteins exposed on the bacterial surface.
FIG. 5.
Analysis of immunological cross-reactivity between Blr and Slr. (A and B) Cross-reactivity of purified proteins. Microtiter wells were coated with Blr (coat Blr) or Slr (coat Slr), and binding of anti-Blr and anti-Slr, diluted as indicated, was analyzed by ELISA. The two antisera were adjusted to have the same titer against the homologous antigen. Preimmune serum was used as a control. (C and D) Cross-reactivity between Blr and Slr expressed on the surface of bacteria. The analysis in panel C employed the S. agalactiae strain Rm69-16, which lacks both capsule and Rib, and the analysis in panel D employed S. pyogenes strain SAM2, which lacks both M protein and protein F. For each bacterial strain, a series of identical samples were mixed with antisera and diluted as indicated, and bound antibodies were detected as described in Materials and Methods. Preimmune serum was used as a control. Each experiment was performed three times with triplicate samples, and each panel shows representative data from one experiment.
Role of Blr in bacterial virulence.
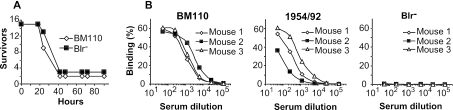
In the S. pyogenes system, studies with a bacterial mutant suggested that Slr contributes to virulence in a mouse model of i.p. infection (46). In contrast, InlA is not a virulence factor for L. monocytogenes in the mouse system, because the interaction of InlA with E-cadherin is species specific (26, 27). To analyze the role of Blr in virulence, we compared the wild-type strain S. agalactiae BM110 with its nonpolar blr mutant and found that both strains were equally virulent in the mouse model of i.p. infection (Fig. 6A). Thus, Blr was not a virulence factor in this model. However, Blr was immunogenic during the course of an infection, as also reported for Slr (28, 46). This conclusion is based on analysis of antisera from mice infected with sublethal doses of different S. agalactiae strains. Mice infected with the type III strain BM110 and the type II strain 1954/92 produced antibodies to Blr (Fig. 6B, left and middle panels), while no antibodies were found in control mice infected with the Blr-negative mutant of BM110 (Fig. 6B, right panel).
FIG. 6.
Blr does not affect virulence in an i.p. infection model but is immunogenic in vivo. (A) Mice in one group were infected i.p. with a 90% lethal dose of S. agalactiae strain BM110, and mice in another group were infected with the same number of bacteria of the nonpolar Blr-negative (Blr−) BM110 mutant, Δblr-36. Each group comprised 15 mice. Deaths were recorded regularly for 96 h. (B) Immune response to Blr in mice infected with a sublethal dose of S. agalactiae. An immune response was detected in mice infected with the Blr-expressing strains BM110 (serotype III) and 1954/92 (serotype II) but not in control mice infected with the Blr-negative BM110 mutant, Δblr-36.
DISCUSSION
Proteins containing LRRs are common among both prokaryotes and eukaryotes. Their functions are diverse, ranging from RNase inhibition to lipopolysaccharide binding, but the common theme is believed to be involvement in different ligand recognitions (23, 25). The crystal structure of an LRR protein was first determined for porcine RNase inhibitor (22), which has a horseshoe-shaped structure in which the concave side is responsible for binding of the ligand, RNase A (24). A number of subsequent structural studies of other LRR proteins have shown great overall similarity to that of the RNase inhibitor. The remarkable ability of LRR proteins to specifically bind different ligands is underlined by the recent identification, in jawless vertebrates, of variable lymphocyte receptors composed of highly diverse LRRs (38, 39).
Here, we have characterized and compared two LRR proteins expressed by important human pathogens, the Blr protein of S. agalactiae and the previously described Slr protein of S. pyogenes (46). Together, these proteins define a family of streptococcal proteins with LRRs. Importantly, the genes for Blr and Slr are present in most, if not all, strains of S. agalactiae and S. pyogenes, respectively, making them of general interest for analysis of streptococcal pathogenesis. The Blr and Slr proteins are predicted to be lipoproteins that are attached to the cell membrane via a cysteine residue in the N-terminal region. Both proteins were indeed shown to be surface exposed, but our data indicate that they are largely camouflaged by other components of the cell envelope, as judged by the poor binding of anti-Blr and anti-Slr antibodies to wild-type bacteria. In S. agalactiae, this effect was mediated by the capsule and, to some extent, the major surface protein Rib, while M protein and protein F caused camouflaging of Slr in S. pyogenes. The demonstration that these surface components mask Blr or Slr does not exclude that other wall components not studied here may also be important for the camouflaging.
The LRR regions of Blr and Slr show considerable residue identity with the LRR region of L. monocytogenes InlA, suggesting that these three surface proteins may have related but not necessarily identical functions. Some support for this hypothesis comes from the fact that the repeats of the three proteins not only exhibit sequence similarity but have similar overall arrangements. In particular, the LRRs have the same length, 22 residues, in each of the three proteins, except for one LRR that has a length of only 21 residues. In Blr and Slr, it is the seventh and fifth LRRs, respectively, that are shorter, and in InlA it is the sixth repeat that is 1 residue shorter. Among the internalins of L. monocytogenes, InlA is unique in having an LRR with 21 residues (49), increasing the significance of this similarity between InlA and Blr/Slr. In InlA, the 21-residue LRR functions as a hinge in which the lack of one amino acid creates a hydrophobic pocket that is essential for the interaction with human E-cadherin (26, 27, 49). It seems possible that also the streptococcal proteins promote adhesion to host tissue and contribute to virulence, although lack of Blr did not affect bacterial virulence in a mouse infection model. Indeed, the negative result in the mouse model does not exclude that Blr contributes to virulence, because the available infection model bypasses the important adhesion step (29) and because Blr might show species-specific binding, like InlA (27).
The accessibility of Blr on the surface of S. agalactiae was increased ∼100-fold by the absence of capsule, and the accessibility of Slr on S. pyogenes was increased >20-fold by the absence of M protein or protein F. Thus, Blr and Slr are efficiently camouflaged by other structures in the cell envelope, although a weak but significant surface exposure could be detected also on wild-type bacteria. To our knowledge, such a dramatic masking effect has not previously been observed for surface proteins of gram-positive bacteria, and its remarkable efficiency suggests that it is of biological significance. For S. agalactiae, it seems possible that the capsule is downregulated during certain phases of an infection, causing exposure of Blr and enabling optimal interactions with the infected host. Observations supporting this hypothesis come from studies showing that acapsular S. agalactiae mutants are more adherent than the parent strains (18, 57). Moreover, the amount of S. agalactiae capsule produced has been correlated with growth rate (40), suggesting that the thickness of the capsule also varies during different growth phases in vivo. Studies of several other bacterial species have also indicated that polysaccharide capsules interfere with the function of different surface structures but are modulated in response to growth phase or environmental signals (15, 48, 53). Interestingly, the capsule did not camouflage the Rib protein, indicating that the capsule of S. agalactiae does not reduce the accessibility of all surface proteins. Possibly, camouflaging by the capsule selectively affects lipoproteins, which are attached to the bacterial cell membrane, and not LPXTG proteins like Rib, which are attached to the peptidoglycan layer (37).
While our studies with S. agalactiae and the published studies with different bacterial systems focus interest on bacterial capsules in regulating the exposure of surface proteins, the ability of other surface proteins to camouflage Slr in S. pyogenes was more surprising. The mechanisms involved remain unclear, but there is precedence from E. coli that protein fimbriae may reduce the accessibility of a surface protein implicated in adhesion (17). Interestingly, expression of M protein and protein F in S. pyogenes is known to be subject to environmental control (5, 31, 59), suggesting that surface exposure of Slr may be enhanced during certain stages of an infection.
Importantly, the camouflaging of a surface protein by other surface structures may cause that protein to go undetected when antibodies are used to analyze surface expression on wild-type bacteria grown in vitro. Indeed, analysis of an S. agalactiae type V strain with antibodies to a large number of putative surface proteins led to the conclusion that many of these proteins, including Blr, were not surface exposed (58). However, it now seems possible that not only Blr but also other proteins were camouflaged by the capsule of that strain. Thus, it will be of interest to include a capsule-negative mutant in future studies of putative S. agalactiae surface proteins. Similarly, it will be of interest to study surface exposure of S. pyogenes proteins not only with wild-type bacteria but also with mutants lacking M protein and/or protein F. Camouflaging may also be of relevance to the recent report that two surface proteins of S. pneumoniae, PsaA and PpmA, were poorly accessible to antibodies (14).
A high degree of cross-reactivity was seen when the purified Blr and Slr proteins were analyzed by ELISA, but the cross-reactivity was unexpectedly found to be very limited when the two proteins were expressed on the surface of bacteria, i.e., under more physiological conditions. Thus, the epitopes that exhibited cross-reactivity in ELISA, when the proteins were exposed on the surface of microtiter wells, were largely inaccessible on the bacterial surface. The low cross-reactivity of the surface-exposed parts (which most likely include the LRR regions) may reflect structural differences that have been selected for during evolution, either because the surface-exposed parts have different functions or because they have evolved to escape cross-reacting antibodies. This result stresses the importance of analyzing cross-reactivity under physiological conditions.
A recent study described the S. agalactiae protein LrrG, which was reported to have LRR-like repeats and was predicted to be attached to the bacterial cell wall via an LPXTG motif (50). This protein is not related to the Blr and Slr proteins studied here, and the repeats in LrrG lack the classical LRR consensus sequence LXXLXLXXNXL. However, LrrG is related to a group of proteins with repeats designated TpLRR, which have been identified for several bacterial pathogens (21, 51). Of note, the LrrG protein is one of the proteins which Tettelin et al. (58) concluded is not exposed on the surface of S. agalactiae, but LrrG was nevertheless reported to be a target for antibodies that protect against S. agalactiae infection (50). Possibly, LrrG is exposed on the bacterial surface during certain stages of an infection but camouflaged by the capsule on bacteria grown in vitro.
In summary, our data show that the Blr protein of S. agalactiae and the previously described Slr protein of S. pyogenes (46) are members of a family of streptococcal LRR proteins that exhibit extensive residue identity but are immunologically diverse. These proteins are associated with the bacterial surface, but on bacteria grown under standard conditions, Blr and Slr are largely camouflaged by other surface components. These interactions provide evidence for a heretofore-unexpected complexity in the relationship between different streptococcal surface structures. It will be of interest to analyze to what extent Blr and Slr are exposed during infections, to evaluate them as possible vaccine components, and to analyze whether they act as adhesins, like the InlA protein of L. monocytogenes.
Acknowledgments
We thank Caroline Olsson and Margaretha Stålhammar-Carlemalm for valuable help and Fredric Carlsson for discussions and a critical reading of the manuscript. Bacterial mutants and/or antisera were kindly provided by Richard Facklam, Emanuel Hanski, Dennis Kasper, Craig Rubens, June Scott, and Michael Wessels.
This work was supported by grants from the Swedish Research Council, Lund University Hospital, The Meningitis Research Foundation (United Kingdom), The Royal Physiographic Society in Lund, and the Trusts of Cornell, Crafoord, Golje, Kock, Lundström, Ronald McDonald, and Österlund.
REFERENCES
- 1.Adamou, J. E., J. H. Heinrichs, A. L. Erwin, W. Walsh, T. Gayle, M. Dormitzer, R. Dagan, Y. A. Brewah, P. Barren, R. Lathigra, S. Langermann, S. Koenig, and S. Johnson. 2001. Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect. Immun. 69:949-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berggård, K., E. Johnsson, E. Morfeldt, J. Persson, M. Stålhammar-Carlemalm, and G. Lindahl. 2001. Binding of human C4BP to the hypervariable region of M protein: a molecular mechanism of phagocytosis resistance in Streptococcus pyogenes. Mol. Microbiol. 42:539-551. [DOI] [PubMed] [Google Scholar]
- 3.Bessen, D. E., and A. Kalia. 2002. Genomic localization of a T serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infect. Immun. 70:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierne, H., S. K. Mazmanian, M. Trost, M. G. Pucciarelli, G. Liu, P. Dehoux, L. Jansch, F. Garcia-del Portillo, O. Schneewind, and P. Cossart. 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43:869-881. [DOI] [PubMed] [Google Scholar]
- 5.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dussurget, O., J. Pizarro-Cerda, and P. Cossart. 2004. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58:587-610. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, M. S., and C. J. Baker. 2001. Group B streptococcal infections, p. 1091-1155. In S. Remington and J. O. Klein (ed.), Infectious disease of the fetus and the newborn infant. W.B. Saunders Company, Philadelphia, Pa.
- 8.Evdokimov, A. G., D. E. Anderson, K. M. Routzahn, and D. S. Waugh. 2001. Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: a leucine-rich repeat protein with the shortest repeating unit. J. Mol. Biol. 312:807-821. [DOI] [PubMed] [Google Scholar]
- 9.Fischetti, V. A. 1989. Streptococcal M protein: molecular design and biological behavior. Clin. Microbiol. Rev. 2:285-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaillard, J. L., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 11.Girardin, S. E., P. J. Sansonetti, and D. J. Philpott. 2002. Intracellular vs extracellular recognition of pathogens—common concepts in mammals and flies. Trends Microbiol. 10:193-199. [DOI] [PubMed] [Google Scholar]
- 12.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 13.Gomez, A., D. Ramon, and P. Sanz. 1994. The Bacillus subtilis lipoprotein LplA causes cell lysis when expressed in Escherichia coli. Microbiology 140:1839-1845. [DOI] [PubMed] [Google Scholar]
- 14.Gor, D. O., X. Ding, D. E. Briles, M. R. Jacobs, and N. S. Greenspan. 2005. Relationship between surface accessibility for PpmA, PsaA, and PspA and antibody-mediated immunity to systemic infection by Streptococcus pneumoniae. Infect. Immun. 73:1304-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerschmidt, S., S. Wolff, A. Hocke, S. Rosseau, E. Muller, and M. Rohde. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect. Immun. 73:4653-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 89:6172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasman, H., T. Chakraborty, and P. Klemm. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J. Bacteriol. 181:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulse, M. L., S. Smith, E. Y. Chi, A. Pham, and C. E. Rubens. 1993. Effect of type III group B streptococcal capsular polysaccharide on invasion of respiratory epithelial cells. Infect. Immun. 61:4835-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inohara, N., and G. Nunez. 2003. NODs: intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 20.Johnsson, E., K. Berggård, H. Kotarsky, J. Hellwage, P. F. Zipfel, U. Sjöbring, and G. Lindahl. 1998. Role of the hypervariable region in streptococcal M proteins: binding of a human complement inhibitor. J. Immunol. 161:4894-4901. [PubMed] [Google Scholar]
- 21.Kajava, A. V., and B. Kobe. 2002. Assessment of the ability to model proteins with leucine-rich repeats in light of the latest structural information. Protein Sci. 11:1082-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobe, B., and J. Deisenhofer. 1993. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature 366:751-756. [DOI] [PubMed] [Google Scholar]
- 23.Kobe, B., and J. Deisenhofer. 1994. The leucine-rich repeat: a versatile binding motif. Trends Biochem. Sci. 19:415-421. [DOI] [PubMed] [Google Scholar]
- 24.Kobe, B., and J. Deisenhofer. 1995. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374:183-186. [DOI] [PubMed] [Google Scholar]
- 25.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725-732. [DOI] [PubMed] [Google Scholar]
- 26.Lecuit, M., S. Dramsi, C. Gottardi, M. Fedor-Chaiken, B. Gumbiner, and P. Cossart. 1999. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18:3956-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lecuit, M., S. Vandormael-Pournin, J. Lefort, M. Huerre, P. Gounon, C. Dupuy, C. Babinet, and P. Cossart. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722-1725. [DOI] [PubMed] [Google Scholar]
- 28.Lei, B., M. Liu, G. L. Chesney, and J. M. Musser. 2004. Identification of new candidate vaccine antigens made by Streptococcus pyogenes: purification and characterization of 16 putative extracellular lipoproteins. J. Infect. Dis. 189:79-89. [DOI] [PubMed] [Google Scholar]
- 29.Lindahl, G., M. Stålhammar-Carlemalm, and T. Areschoug. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18:102-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinon, F., and J. Tschopp. 2004. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell 117:561-574. [DOI] [PubMed] [Google Scholar]
- 31.McIver, K. S., A. S. Heath, and J. R. Scott. 1995. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 63:4540-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mengaud, J., H. Ohayon, P. Gounon, R. M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84:923-932. [DOI] [PubMed] [Google Scholar]
- 33.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 34.Miller, L., L. Gray, E. Beachey, and M. Kehoe. 1988. Antigenic variation among group A streptococcal M proteins. Nucleotide sequence of the serotype 5 M protein gene and its relationship with genes encoding types 6 and 24 M proteins. J. Biol. Chem. 263:5668-5673. [PubMed] [Google Scholar]
- 35.Musser, J. M., S. J. Mattingly, R. Quentin, A. Goudeau, and R. K. Selander. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B streptococcus) causing invasive neonatal disease. Proc. Natl. Acad. Sci. USA 86:4731-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakata, M., A. Podbielski, and B. Kreikemeyer. 2005. MsmR, a specific positive regulator of the Streptococcus pyogenes FCT pathogenicity region and cytolysin-mediated translocation system genes. Mol. Microbiol. 57:786-803. [DOI] [PubMed] [Google Scholar]
- 37.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pancer, Z., C. T. Amemiya, G. R. Ehrhardt, J. Ceitlin, G. L. Gartland, and M. D. Cooper. 2004. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 430:174-180. [DOI] [PubMed] [Google Scholar]
- 39.Pancer, Z., N. R. Saha, J. Kasamatsu, T. Suzuki, C. T. Amemiya, M. Kasahara, and M. D. Cooper. 2005. Variable lymphocyte receptors in hagfish. Proc. Natl. Acad. Sci. USA 102:9224-9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paoletti, L. C., R. A. Ross, and K. D. Johnson. 1996. Cell growth rate regulates expression of group B Streptococcus type III capsular polysaccharide. Infect. Immun. 64:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Casal, J., J. A. Price, E. Maguin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809-819. [DOI] [PubMed] [Google Scholar]
- 43.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lutticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 44.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 45.Reid, S. D., N. M. Green, J. K. Buss, B. Lei, and J. M. Musser. 2001. Multilocus analysis of extracellular putative virulence proteins made by group A streptococcus: population genetics, human serologic response, and gene transcription. Proc. Natl. Acad. Sci. USA 98:7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reid, S. D., A. G. Montgomery, J. M. Voyich, F. R. DeLeo, B. Lei, R. M. Ireland, N. M. Green, M. Liu, S. Lukomski, and J. M. Musser. 2003. Characterization of an extracellular virulence factor made by group A streptococcus with homology to the Listeria monocytogenes internalin family of proteins. Infect. Immun. 71:7043-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubens, C. E., L. M. Heggen, R. F. Haft, and M. R. Wessels. 1993. Identification of cpsD, a gene essential for type III capsule expression in group B streptococci. Mol. Microbiol. 8:843-855. [DOI] [PubMed] [Google Scholar]
- 48.Schembri, M. A., D. Dalsgaard, and P. Klemm. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 186:1249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schubert, W. D., C. Urbanke, T. Ziehm, V. Beier, M. P. Machner, E. Domann, J. Wehland, T. Chakraborty, and D. W. Heinz. 2002. Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 111:825-836. [DOI] [PubMed] [Google Scholar]
- 50.Seepersaud, R., S. B. Hanniffy, P. Mayne, P. Sizer, R. Le Page, and J. M. Wells. 2005. Characterization of a novel leucine-rich repeat protein antigen from group B streptococci that elicits protective immunity. Infect. Immun. 73:1671-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shevchenko, D. V., D. R. Akins, E. Robinson, M. Li, T. G. Popova, D. L. Cox, and J. D. Radolf. 1997. Molecular characterization and cellular localization of TpLRR, a processed leucine-rich repeat protein of Treponema pallidum, the syphilis spirochete. J. Bacteriol. 179:3188-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stålhammar-Carlemalm, M., L. Stenberg, and G. Lindahl. 1993. Protein Rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J. Exp. Med. 177:1593-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.St. Geme, J. W., III, and D. Cutter. 1996. Influence of pili, fibrils, and capsule on in vitro adherence by Haemophilus influenzae type b. Mol. Microbiol. 21:21-31. [DOI] [PubMed] [Google Scholar]
- 54.Sutcliffe, I. C., and D. J. Harrington. 2004. Putative lipoproteins of Streptococcus agalactiae identified by bioinformatic genome analysis. Antonie Leeuwenhoek 85:305-315. [DOI] [PubMed] [Google Scholar]
- 55.Sutcliffe, I. C., and R. R. Russell. 1995. Lipoproteins of gram-positive bacteria. J. Bacteriol. 177:1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talay, S. R., P. Valentin-Weigand, P. G. Jerlstrom, K. N. Timmis, and G. S. Chhatwal. 1992. Fibronectin-binding protein of Streptococcus pyogenes: sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infect. Immun. 60:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura, G. S., J. M. Kuypers, S. Smith, H. Raff, and C. E. Rubens. 1994. Adherence of group B streptococci to cultured epithelial cells: roles of environmental factors and bacterial surface components. Infect. Immun. 62:2450-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.VanHeyningen, T., G. Fogg, D. Yates, E. Hanski, and M. Caparon. 1993. Adherence and fibronectin binding are environmentally regulated in the group A streptococci. Mol. Microbiol. 9:1213-1222. [DOI] [PubMed] [Google Scholar]
- 60.Venkatesan, M. M., J. M. Buysse, and A. B. Hartman. 1991. Sequence variation in two ipaH genes of Shigella flexneri 5 and homology to the LRG-like family of proteins. Mol. Microbiol. 5:2435-2445. [DOI] [PubMed] [Google Scholar]
- 61.Wästfelt, M., M. Stålhammar-Carlemalm, A. M. Delisse, T. Cabezon, and G. Lindahl. 1996. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J. Biol. Chem. 271:18892-18897. [DOI] [PubMed] [Google Scholar]
- 62.Wessels, M. R., A. E. Moses, J. B. Goldberg, and T. J. DiCesare. 1991. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc. Natl. Acad. Sci. USA 88:8317-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wessels, M. R., J. B. Goldberg, A. E. Moses, and T. J. DiCesare. 1994. Effects on virulence of mutations in a locus essential for hyaluronic acid capsule expression in group A streptococci. Infect. Immun. 62:433-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y., A. W. Masi, V. Barniak, K. Mountzouros, M. K. Hostetter, and B. A. Green. 2001. Recombinant PhpA protein, a unique histidine motif-containing protein from Streptococcus pneumoniae, protects mice against intranasal pneumococcal challenge. Infect. Immun. 69:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]