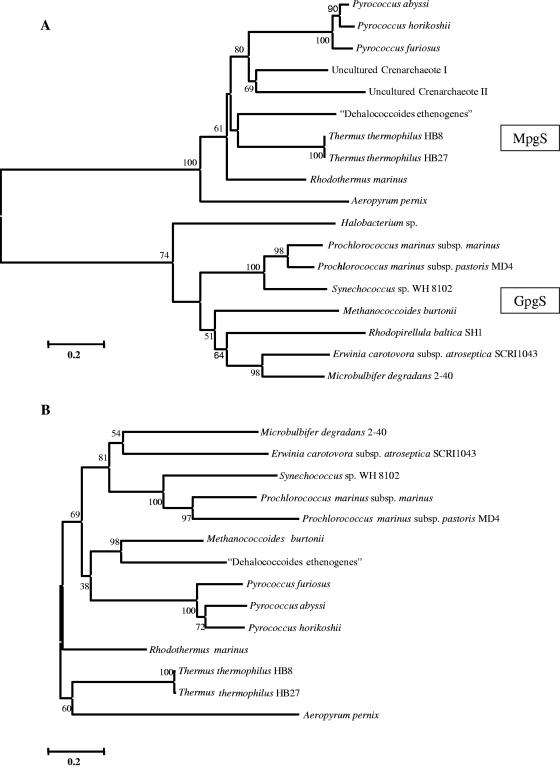
FIG. 4.
Unrooted phylogenetic trees based on available amino acid sequences of GpgS, MpgS, GpgS, and MpgS homologues (A) and GpgP, MpgP, and GpgP homologues (B). The Clustal X program (40) was used for sequence alignment, and the MEGA2 program (24) was used to generate the phylogenetic tree. The significance of the branching order was evaluated by bootstrap analysis of 1,000 computer-generated trees. The bootstrap values are indicated. Bar, 0.2 changes/site.