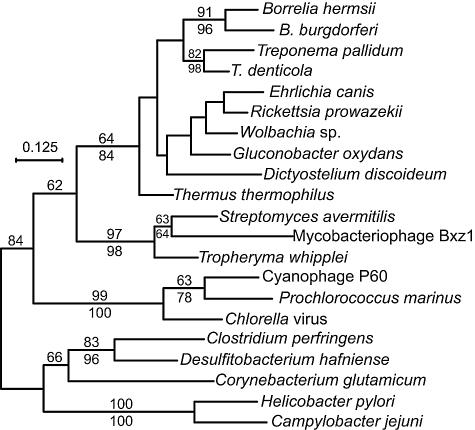
FIG. 3.
Phylogram of codon-aligned nucleotide sequences of thyX genes from the following organisms (with accession numbers): Borrelia burgdorferi (NC_001857), B. hermsii (AY623744), Campylobacter jejuni (NC_003912), Chlorella virus (NC_000852), Clostridium perfringens (BA000016), Corynebacterium glutamicum (BX927153), cyanophage P60 (AF338467), Desulfitobacterium hafniense (NZ_AAAW03000212), Dictyostelium discoideum (M27713), Ehrlichia canis (NZ_AAEJ01000001), Gluconobacter oxydans (NC_006677), Helicobacter pylori (NC_000921), mycobacteriophage Bxz1 (AY129337), Prochlorococcus marinus (AE017161), Rickettsia prowazekii (NC_000963), Streptomyces avermitilis (BA000030), Thermus thermophilus (NC_005835), Treponema denticola (NC_002967), T. pallidum (AE001268), Tropheryma whipplei (AE016852), and a Wolbachia endosymbiont of Drosophila melanogaster (NC_002978). The third position of each codon and positions with gaps were excluded from the analysis. Nodes with bootstrap values with >61% support by maximum likelihood (100 replicates; shown above the line) or neighbor-joining (1,000 replicates; shown below the line) distance criteria are indicated. Bar, nucleotide distance.