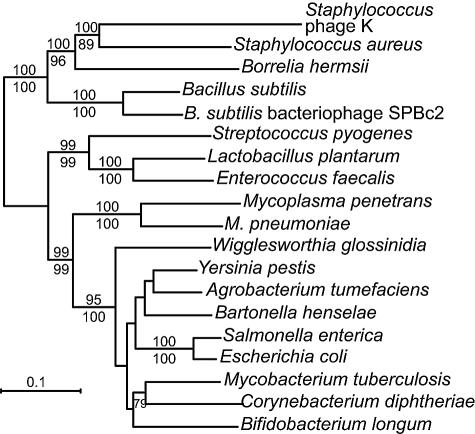
FIG. 4.
Phylogram of codon-aligned concatenated nucleotide sequences of nrdE and nrdF genes from the following bacteria and viruses (with accession numbers): Agrobacterium tumefaciens (AE008981), Bacillus subtilis (NC_000964), B. subtilis bacteriophage SPBc2 (AF020713), Bartonella henselae (BX897699), Bifidobacterium longum (AE014295), Borrelia hermsii (AY623744), Corynebacterium diphtheriae (NC_002935), Enterococcus faecalis (NC_004668), Escherichia coli (U00096), Lactobacillus plantarum (NC_004567), Mycobacterium tuberculosis (NC_000962), Mycoplasma penetrans (BA000026), Mycoplasma pneumoniae (AE000050), Salmonella enterica (NC_006511), Staphylococcus aureus (NC_002758), Staphylococcus phage K (NC_005880), Streptococcus pyogenes (AE010059), Wigglesworthia glossinidia (BA000021), and Yersinia pestis (AE017136). Nucleotide positions with gaps and the third base of each codon in the aligned sequences were excluded from the analysis. Bootstrap values of nodes with >71% support by maximum likelihood (100 replicates; shown above the line) or neighbor-joining (1,000 replicates; shown below the line) distance criteria are indicated. Bar, nucleotide distance.