Abstract
Mga, or the multigene regulator of the group A streptococcus (GAS) (Streptococcus pyogenes), is a transcriptional regulator of virulence genes important for colonization and immune evasion. All serotypes of the GAS possess one of two divergent mga alleles (mga-1 or mga-2), and orthologues of Mga have also been identified in other pathogenic streptococci. To date, the only functional motifs established within Mga are two amino-terminal DNA-binding domains (HTH-3 and HTH-4). To uncover novel domains, a random mutagenesis screen using an M6 Mga (mga-1) was undertaken to find mutations leading to a defect in transcriptional activation of the Mga-regulated emm gene. In addition to mutations in the established DNA-binding domains, the screen also revealed mutations in a region conserved among several Mga orthologues. Alanine scanning helped resolve the boundaries of this conserved Mga domain (CMD-1) spanning from residues 10 to 15 of the protein, with the two flanking amino acid residues likely involved in protein stability. Transcriptional reporter analyses demonstrated the importance of CMD-1 for activation of Pemm and autoactivation of Pmga in the serotype M6 Mga. Mutational analyses showed that both CMD-1 and HTH-4 are also necessary for activation of the promoter target Pmrp in a divergent serotype M4 Mga (mga-2), suggesting a conserved functionality. However, in contrast to M6, the M4 Mga mutants did not show a defect in autoregulation. Mutation of similar conserved residues in the Mga-like regulator DmgB from S. dysgalactiae subsp. dysgalactiae showed that CMD-1 and HTH-4 are critical for transcriptional activation in this orthologue, implying that a common mechanism of virulence gene activation may exist for members of the Mga family of regulators.
The genus Streptococcus is a large heterogeneous collection of gram-positive cocci, with each species differing in its requirements for optimum growth and demonstrating a defining hemolytic reaction on blood agar plates. One unifying theme of this genus is that many of its members colonize a broad range of tissue sites in humans and/or animals (6). A clear example is the group A streptococcus (GAS) (Streptococcus pyogenes), which is able to cause a wide array of diseases throughout the human body ranging from self-limiting to life-threatening. Understanding how pathogenic streptococci survive and elicit disease throughout their host remains an underlying goal of research on these organisms.
Survival within different niches stems from the repertoire of virulence genes these pathogens possess and the ability to regulate their expression in response to specific environmental conditions. Differential gene expression in the GAS and other streptococci is often controlled by two-component signal transduction systems, which allow the bacteria to sense their environment and respond via activation or inactivation of the appropriate transcriptional regulators. However, not all transcriptional regulators that respond to environmental stimuli are part of an established two-component system. For example, the GAS contain several “stand-alone” response regulators, which are defined as environmentally responsive transcriptional regulators that contain no known sensory components (13).
One such stand-alone response regulator is Mga, the multigene regulator of the GAS. Although the mechanism by which Mga can respond to environmental signals remains undefined, it has been shown to positively activate its own expression (18, 19) along with the expression of other genes within its regulon in response to increased CO2 levels, body temperature, and exponential-phase growth (4, 15, 17, 22). In the GAS, the mga regulon is composed of genes important for colonization and immune evasion, such as those encoding the antiphagocytic M protein (emm), an immunoglobulin-binding protein (mrp), a collagen-like protein (scl), and a C5a peptidase (scpA).
While all strains of the GAS contain an mga gene (3), the architecture of the mga regulon differs from strain to strain in both gene composition and arrangement (11, 29). Furthermore, two mga alleles have been described within the GAS (mga-1 and mga-2) based upon the ability to hybridize to an oligonucleotide probe (12) and are associated with different gene patterns at the mga locus and tissue tropism of the serotype (11). The Mga proteins produced from the two alleles are most divergent within the C-terminal end of the protein (2), showing a maximal amino acid divergence of 20.7% (3). Despite the divergence, both mga alleles have been shown to be functionally equivalent in an mga-1 deletion strain (2).
S. pyogenes is not the only streptococcal species that appears to contain a gene analogous to mga. Geyer and Schmidt identified an orthologue in S. dysgalactiae subsp. equisimilis named mgc (8) for the multigene regulator of the group C streptococcus (GCS). The Mgc protein is 51% identical and 64% similar to the Mga (mga-1) from the serotype M6 GAS strain D471 (8). Another GCS orthologue designated dmgB, exhibiting 45% identity and 61% similarity to Mga, has been identified in S. dysgalactiae subsp. dysgalactiae (28). The mgc and dmgB loci closely resemble that of the GAS mga in that a gene encoding an M protein homolog (emm and demB, respectively) lies directly downstream of the aforementioned GCS genes. Recently, a virulence regulator showing 51% similarity and 25% identity to Mga from the GAS was found in S. pneumoniae, called mgrA for the Mga-like repressor A (9, 10). In contrast to Mga and DmgB/Mgc, MgrA appears to repress transcription of genes in the unlinked rlrA pathogenicity islet in the S. pneumoniae TIGR4 genome (10). Additional predicted regulators showing similarity to Mga can be found in the genome sequences of other streptococcal species, including S. equi, S. gordonii, and S. mitis.
Currently, only Mga has been extensively characterized and serves as an excellent model system for this family of Mga-like virulence regulators. A serotype M6 Mga (mga-1) is able to activate transcription by interacting with specific DNA sites within regulated (Pemm, PscpA, and PsclA) and autoregulated (Pmga) promoters (1, 14, 18). Two amino-terminal helix-turn-helix (HTH) domains necessary for DNA binding and transcriptional activation were found in M6 Mga: one (HTH-4) that is absolutely essential for binding to all targets and a second (HTH-3) that serves an accessory role, primarily in autoregulation from Pmga (16). In addition, two amino acids involved in transcriptional activation of a divergent Mga (mga-2) have been identified (27). Finally, two putative response regulator receiver domains have been suggested to exist in Mga based on homology to other two-component systems (20); however, no molecular evidence for these domains has been presented.
Since little is known about how Mga is able to sense environmental stimuli and activate virulence gene transcription, a genetic screen was devised to uncover functional domains within a serotype M6 Mga (mga-1) important for transcriptional activation. Using this approach, we have identified a novel conserved domain that is important for activation of downstream target genes not only in divergent Mga proteins from the GAS, but also for the orthologue DmgB from GCS. Our data suggest that differences exist between divergent Mga proteins and their contribution to autoregulation. Finally, we have shown that HTH-4 is necessary for transcriptional activation in two members of the Mga family of virulence regulators.
MATERIALS AND METHODS
Bacterial strains and media.
S. pyogenes JRS519 is a Δmga kanamycin-resistant derivative of the serotype M6 strain JRS4 (17, 24). The S. pyogenes vectors for integration (VIT) strain RTG229 is a derivative of JRS4 (7). Both the M6 Pemm-gusA GAS reporter strain KSM148.174 and the M6 Pmrp-gusA GAS reporter strain KSM149 are unmarked mga deletion (Δmga) derivatives of the VIT strain RTG229 (27). The clinical isolate AP4 is a serotype M4 GAS strain (25). Escherichia coli DH5α (hsdR17 recA1 gyrA endA1 relA1) was used as the host for all plasmid constructions, and protein purifications used the E. coli strain BL21(DE3) containing the T7 RNA polymerase (26).
GAS strains were grown in Todd-Hewitt medium supplemented with 0.2% yeast extract (THY), while E. coli strains were grown in Luria-Bertani broth (LB). Growth of GAS was measured on a Klett-Summerson photoelectric colorimeter using an A filter. The following concentrations of antibiotics were used: ampicillin at 100 μg/ml for E. coli; spectinomycin at 100 μg/ml for both E. coli and GAS; and erythromycin at 500 μg/ml for E. coli and 1 μg/ml for GAS.
DNA manipulation.
Plasmid DNA was isolated from E. coli using the Wizard miniprep kit (Promega) or from the GAS as described below. Genomic DNA was isolated from the GAS using the FastDNA prep and a FastPrep cell disruptor (Bio 101). DNA fragments were purified from agarose gels using the QIAquick gel extraction kit (QIAGEN). All PCR, with the exception of that used to generate random mutations, was performed using Pfu Turbo DNA polymerase (Stratagene) and resulting products were purified with the QIAquick PCR purification system (QIAGEN). All site-specific mutations were generated with the QuickChange site-directed mutagenesis kit (Stratagene), using mutagenic oligonucleotides synthesized by Integrated DNA Technologies. DNA sequencing was done by the McDermott Center sequencing core facility at the University of Texas Southwestern Medical Center.
Isolation of plasmid DNA from the GAS.
Cells from a 50-ml overnight culture were resuspended in P1 solution (QIAGEN) containing 20 mg/ml lysozyme, mixed, and incubated for 30 min at 37°C. Samples were lysed with the FastPrep cell disruptor (Bio 101) and allowed to settle and the supernatant was mixed with 300 μl of a 1% sodium dodecyl sulfate-0.2 M NaOH solution. After 5 min at room temperature, 300 μl of 2.5 M potassium acetate, pH 4.5, was added, mixed by inversion, and centrifuged (5 min, 13,000 rpm). The supernatant was applied to a miniprep column (QIAGEN) and plasmid DNA was eluted using the manufacturer's protocol.
Random PCR mutagenesis screen.
Mutagenic PCR was performed across the 5′ region of mga using the GeneMorph PCR mutagenesis kit (Stratagene) on plasmid pKSM162 (16) with primers RMut1-R2 and Mga6-150 (Table 1) and an initial amount of DNA of 0.110 μg. The resulting PCR product was digested with AvaI and SpeI, ligated with AvaI- and SpeI-digested pKSM318 (27), transformed into E. coli DH5α, and grown overnight in 10 ml LB broth containing spectinomycin. A similar procedure was performed for the 3′ portion of mga except primers OYR-29 and RMut2-L were used on various concentrations of pKSM162 ranging from 0.0552 to 0.838 μg. The reduction in starting concentration was to compensate for the increased size of the 3′ fragment as per the manufacturer's protocol. Also, both the PCR product and the vector pKSM318 were digested with SpeI and SphI for this fragment.
TABLE 1.
PCR primers and their relevant targetsa
| Target and primer | Sequence | Reference |
|---|---|---|
| 23S rRNA | ||
| rRNA-23SL | GGAAGGTAAGCCAAAGAGAG | 23 |
| rRNA-23SR | TCCTAGTTGTCTGTGCAACC | 23 |
| Pemm | ||
| Pemm-L1 | GCATGGATCCCATCGCAAAGAGCTTA | 16 |
| Pemm-R1 | GCGGCTCGAGTAGTGTCTATTCGTGTTATT | 16 |
| Pmga | ||
| OYL-25 | TACCATAAAATACCTTTC | 16 |
| OYR-25 | GGTTGTACCATAACAGTC | 16 |
| mga6 | ||
| Mga6-150 | gcgtcaaagcttctaATCTCCTGATACTTGTACGG | This study |
| Mga-Pet1_Nde | ggggcatATGTATGTAAGTAAGTTGTTT | 1 |
| Mga-Pet2_Xho | aactcgagAGTTGTGGAGGGGG | 1 |
| OYR-29 | AAACCAACGCCTATTTGACGCATAC | This study |
| RMut1-R2 | cccgcctcgagAAAGAAGGGTATACAAGG | This study |
| RMut2-L | TCGACCTGCAGgcatgcaaa | This study |
| Mutagenic mga6 | ||
| M6 mga Q10R-a | GTTGTTTACAAGTCgACAGTGGAGAGAACTAAAATTAATCTCATACGTAACGG | This study |
| M6 mga Q10R-b | CCGTTACGTATGAGATTAATTTTAGTTCTCTCCACTGTcGACTTGTAAACAAC | This study |
| M6 mga Q11A-a | GTTGTTTACAAGTCAAgcGTGGAGAGAACTAAAATTAATCTCATACGTAACGG | This study |
| M6 mga Q11A-b | CCGTTACGTATGAGATTAATTTTAGTTCTCTCCACgcTTGACTTGTAAACAAC | This study |
| M6 mga W12A-a | GTTGTTTACAAGTCAACAGgcGAGAGAACTAAAATTAATCTCATACGTAACGG | This study |
| M6 mga W12A-b | CCGTTACGTATGAGATTAATTTTAGTTCTCTCgcCTGTTGACTTGTAAACAAC | This study |
| M6 mga W12R-a | GTTGTTTACAAGTCAACAGcGGAGAGAACTAAAATTAATCTCATACGTAACGG | This study |
| M6 mga W12R-b | CCGTTACGTATGAGATTAATTTTAGTTCTCTCCgCTGTTGACTTGTAAACAAC | This study |
| M6 mga R13A-a | GTTTACAAGTCAACAGTGGgcAGAACTAAAATTAATCTCATACGTAACGG | This study |
| M6 mga R13A-b | CCGTTACGTATGAGATTAATTTTAGTTCTgcCCACTGTTGACTTGTAAAC | This study |
| M6 mga E14A-a | GTTGTTTACAAGTCAACAGTGGAGAGcACTAAAATTAATCTCATACGTAACGG | This study |
| M6 mga E14A-b | CCGTTACGTATGAGATTAATTTTAGTgCTCTCCACTGTTGACTTGTAAACAAC | This study |
| M6 mga L15A-a | GTTTACAAGTCAACAGTGGAGAGAAgcAAAATTAATCTCATACGTAACGG | This study |
| M6 mga L15A-b | CCGTTACGTATGAGATTAATTTTgcTTCTCTCCACTGTTGACTTGTAAAC | This study |
| Mutagenic mga4 | ||
| M4 mga Q11R-a | GCATGTAAGTAAATTGTTTACTAGCCAACgATGGAGAGAATTGAAACTG | This study |
| M4 mga Q11R-b | CAGTTTCAATTCTCTCCATcGTTGGCTAGTAAACAATTTACTTACATGC | This study |
| M4 mga R13A-a | GTTTACTAGCCAACAATGGgcAGAATTGAAACTGATTTCATATTTAACAG | This study |
| M4 mga R13A-b | CTGTTAAATATGAAATCAGTTTCAATTCTgcCCATTGTTGGCTAGTAAAC | This study |
| M4 mga HTH4-a | GCTGAAGAGCTGTTTGTCAGCgcAgCTACCCTCAAACGCC | This study |
| M4 mga HTH4-b | GGCGTTTGAGGGTAGcTgcGCTGACAAACAGCTCTTCAGC | This study |
| Mutagenic dmgB | ||
| dmgB Q11R-a | CTCTTTACAACAAAACgGTGGAGAGAATTGGAGCTAATTGCGC | This study |
| dmgB Q11R-b | GCGCAATTAGCTCCAATTCTCTCCACcGTTTTGTTGTAAAGAG | This study |
| dmgB R13A-a | CTCTTTACAACAAAACAGTGGgcAGAATTGGAGCTAATTGCGC | This study |
| dmgB R13A-b | GCGCAATTAGCTCCAATTCTgcCCACTGTTTTGTTGTAAAGAG | This study |
| dmgB L42A-a | GTGTGAGAGATTAAACTGCTCACTCgcAACTTTACAATCATGTG | This study |
| dmgB L42A-b | CACATGATTGTAAAGTTgcGAGTGAGCAGTTTAATCTCTCACAC | This study |
| dmgB HTH4-a | GAGCTGTTTGTCAGCgcGgCAACACTCAAGCGTTTGATTG | This study |
| dmgB HTH4-b | CAATCAAACGCTTGAGTGTTGcCgcGCTGACAAACAGCTC | This study |
Mutagenic or noncomplementary sequences are in lowercase and introduced restriction sites are underlined.
Plasmid DNA was extracted from the entire overnight culture, and 1 μg of plasmid DNA was transformed into the reporter strain KSM148.174 (27). Following selection on THY agar containing spectinomycin and 175 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc) (Gold Biotechnology), GAS transformants that were either light blue or white on plates were subjected to Western analysis to determine protein levels as described below. Plasmid DNA was extracted from the selected clones and amplified via subsequent transformation into E. coli DH5α for DNA sequencing. Sequence alignments were performed with the Vector NTI software (Invitrogen) using the ClustalW algorithm, and genes containing more than two mutations were discarded from further analysis.
Whole-cell GAS protein extracts.
Whole-cell GAS protein extractions were performed as previously described (16). Briefly, mid-logarithmic-phase GAS cultures were harvested by centrifugation and resuspended in saline containing 1× complete protease inhibitor cocktail (Roche). Cells were lysed with the FastPrep cell disruptor (Bio 101) and the soluble fraction was recovered after centrifugation. Total protein concentrations were determined using the protein assay kit (Bio-Rad).
Western blot analyses.
Proteins were analyzed by Western blot as previously described (16). Blots were incubated with either a 1:2,000 dilution of anti-His tag monoclonal antibody (Novagen) or a 1:1,000 dilution anti-Mga-pep2 antiserum (16), then incubated with a 1:25,000 dilution of either anti-mouse (Chemicon) or anti-rat (Santa Cruz Biotechnologies) horseradish peroxidase-conjugated secondary antibody, respectively, and visualized using the Western Lightning chemiluminescence system (Perkin Elmer). For a loading control, blots were stripped in a solution containing 2% sodium dodecyl sulfate, 50 mM dithiothreitol (DTT), and 50 mM Tris-HCl, pH 7.0, for 30 min at 70°C and reprobed with a 1:50,000 dilution of mouse anti-Hsp60 monoclonal antibody (StressGen Biotechnologies Corp.).
GusA reporter assays.
Soluble whole-cell lysates were isolated from the GAS grown to mid-logarithmic phase (65 Klett units) as described above. GusA activity was determined for each lysate (100 μl) as described by Eichenbaum et al. (5). Total protein concentrations were determined as above. GusA units are defined as the optical density at 420 nm (OD420)/total protein concentration of the lysate.
Construction of a Pmga-gusA reporter strain containing a ΔPmga-mga allele.
Pmga-gusA reporter strain KSM231.310 was generated by transforming the mga-deleted M6 VIT strain VIT231 with pPmga-gusA (27) linearized by XmnI, which created a Δmga reporter strain containing a single-copy transcriptional fusion of the native M6 mga promoter to gusA (Pmga-gusA).
Construction of the site-specific Pspac-mga6-his plasmids pQ10R, pQ11A, pW12A, pW12R, pR13A, pE14A, and pL15A.
Site-specific mutants of M6 Mga produced from the constitutive promoter Pspac and containing a carboxy-terminal 6X His tag were generated as described above using the QuickChange site-directed mutagenesis kit (Stratagene), pKSM318 (Table 2) template DNA, and the corresponding mga6 mutagenic oligonucleotides (Table 1), resulting in the different plasmids.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristics | Reference |
|---|---|---|
| pQ10R | M6 mga mutant Q10R under constitutive Pspac | This study |
| pQ11A | M6 mga mutant Q11A under constitutive Pspac | This study |
| pQ11R | M6 mga mutant Q11R under constitutive Pspac | This study |
| pW12A | M6 mga mutant W12A under constitutive Pspac | This study |
| pW12R | M6 mga mutant W12R under constitutive Pspac | This study |
| pR13A | M6 mga mutant R13A under constitutive Pspac | This study |
| pE14A | M6 mga mutant E14A under constitutive Pspac | This study |
| pL15A | M6 mga mutant L15A under constitutive Pspac | This study |
| pKSM152 | S. dysgalactiae locus containing both wild-type dmgB and demB | This study |
| pKSM162 | Wild-type mga6 under constitutive Pspac | 16 |
| pKSM170 | Wild-type M6 mga-his in E. coli expression vector | 1 |
| pKSM318 | Wild-type mga6-his under constitutive Pspac | 27 |
| pKSM318.1H | M6 mga mutant in HTH-1 under constitutive Pspac | This study |
| pKSM318.4H | M6 mga mutant in HTH-4 under constitutive Pspac | This study |
| pKSM322 | Wild-type mga4-his under native Pmga4 | 27 |
| pKSM339 | dmgB mutant Q11R under the native promoter | This study |
| pKSM340 | dmgB mutant R13A under the native promoter | This study |
| pKSM341 | dmgB mutant in HTH-4 under the native promoter | This study |
| pKSM342 | dmgB mutant L42A in HTH-1 under the native promoter | This study |
| pKSM344 | M6 mga-his mutant Q11R in E. coli expression vector | This study |
| pKSM345 | M6 mga-his mutant R13A in E. coli expression vector | This study |
| pKSM346 | M4 mga-his mutant Q11R under native Pmga4 | This study |
| pKSM347 | M4 mga-his mutant R13A under native Pmga4 | This study |
| pKSM348 | M4 mga-his mutant in HTH-4 under native Pmga4 | This study |
| pKSM354 | M6 mga-his mutant W12R in E. coli expression vector | This study |
| pKSM355 | M6 mga-his mutant E14A in E. coli expression vector | This study |
Construction of the control Pspac-mga6-his plasmids pKSM318.1H and pKSM318.4H.
Plasmids that contained mutations to disrupt either the essential DNA binding HTH-4 motif or two nonessential residues of M6 mga under the constitutive Pspac were constructed as follows: the 464-bp HindIII-SpeI fragment containing either the HTH-4 motif or K31A/D32A from pKSM164.4c or pKSM164.1c (16) was cloned into HindIII- and SpeI-digested pKSM318 (27) to produce pKSM318.4H and pKSM318.1H, respectively.
Construction of the Pmga4-mga4-his mutant plasmids pKSM346, pKSM347, and pKSM348.
Site-specific mutants of the class II serotype M4 Mga, Q11R and R13A, were generated as described above using the mutagenic oligonucleotides M4 mga Q11R-a, M4 mga Q11R-b, M4 mga R13A-a, and M4 mga R13A-b (Table 1) in pKSM322 (Table 2), resulting in pKSM346 and pKSM347, respectively. A mutant disrupting the HTH-4 domain, named pKSM348, was also constructed by site-specific mutagenesis of pKSM322 using the mutagenic oligonucleotides M4 mga HTH4-a and M4 mga HTH4-b (Table 1).
Construction of the PdmgB-dmgB mutant plasmids pKSM339, pKSM340, pKSM341, and pKSM342.
Site-specific mutations at positions 11 and 13 were constructed in the Mga orthologue from S. dysgalactiae, DmgB, using the mutagenic oligonucleotides dmgB Q11R-a, dmgB Q11R-b, dmgB R13A-a, and dmgB R13A-b (Table 1) in pKSM152 (Table 2), resulting in pKSM339 and pKSM340, respectively. Control plasmids containing either mutations in the HTH-4 domain, pKSM341, or an arbitrary mutation at position 42, pKSM342, were also constructed by site-specific mutagenesis of pKSM152 using the mutagenic oligonucleotides dmgB HTH4-a, dmgB HTH4-b, dmgB L42A-a, and dmgB L42A-b, respectively (Table 1).
Expression and purification of Mga-His fusion proteins from E. coli.
Plasmids expressing carboxy-terminal 6X His fusions to M6 Mga mutants Q11R, W12R, R13A, and E14A were constructed as follows: a 1.6-kb DNA fragment of the mga coding sequence was amplified from plasmid DNA encoding the corresponding mutant Q11R, W12R, R13A, and E14A using primers Mga-Pet1_Nde and Mga-Pet2_Xho (Table 1). The resulting PCR fragments were purified, digested with NdeI and XhoI, and inserted into NdeI- and XhoI-digested vector pKSM170 (1) to generate the mga-his fusion alleles for purification of Q11R-His (pKSM344), W12R-His (pKSM354), R13A-His (pKSM345), and E14A-His (pKSM355), respectively (Table 2). Mga-His proteins were purified from E. coli as previously described (1).
EMSA.
Promoter probes for Pemm and Pmga were generated by PCR amplification from serotype M6 strain JRS4 genome DNA using the relevant primer pairs, Pemm-L1 and Pemm-R1 and OYR-25 and OYL-25, respectively (Table 1). An electrophoretic mobility shift assay (EMSA) was performed as previously described (14). Briefly, constant amounts of probe end labeled with [γ-32P]ATP were incubated with increasing concentrations of purified Mga-His proteins for 15 min at 16°C before being separated on a 5% polyacrylamide gel.
Northern blot analysis.
Total RNA was isolated from the GAS grown to mid-logarithmic phase using the FastRNA Pro Blue kit and FastPrep cell disruptor (Bio 101). Northern blot analysis was performed using the NorthernMax system (Ambion) as previously described (23). As a loading control, blots were stripped and reprobed with a 23S rRNA probe (Table 1).
Alignment of Mga orthologues.
A sequence alignment of different Mga and Mga-like protein sequences was performed using the ClustalW algorithm in the AlignX module of VectorNTI. The alignment included Mga protein sequences representing mga-1 alleles from GAS serotypes M1 SF370 (GI-15675800) and M6 JRS4 (GI-153733) as well as mga-2 alleles from GAS serotypes M4 AP4 (GI-1246852) and M49 531 (GI-56808536). Protein sequences for S. pneumoniae MgrA (GI-17368568), S. dysgalactiae subsp. equisimilis Mgc (GI-6782393), and S. dysgalactiae subsp. dysgalactiae DmgB (GI-6689248) were also utilized. Additional Mga-like proteins were obtained by BLAST interrogation of the unfinished genomes for S. equi (Sanger Institute), S. mitis (TIGR), and S. gordonii (TIGR). Conserved regions were defined as amino acids with ≥70% identity among homologues and spanned more than two adjacent residues.
RESULTS
Random mutagenesis screen for Mga mutants defective in transcriptional activation.
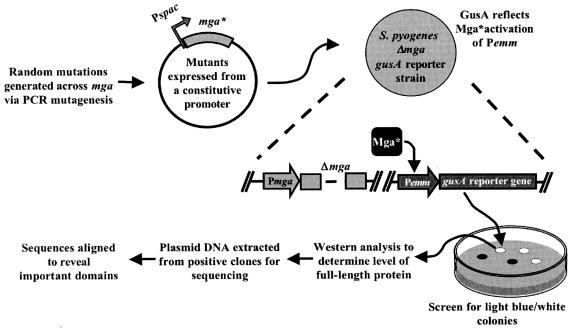
A nonbiased strategy using PCR-generated mutations was devised to identify functional residues within Mga involved in activation of Mga-dependent genes (Fig. 1). PCR amplification using a defective DNA polymerase was utilized to generate random mutations across the 1,590-base-pair mga gene from the serotype M6 strain JRS4 (see Materials and Methods), which was arbitrarily divided into two fragments (N and C terminal) at a unique SpeI site for ease of handling. Mutated mga fragments were then cloned under the constitutive Pspac promoter in plasmid pKSM318 (27) such that transcription levels in the screen would not reflect autoactivation from the native Pmga promoter. The resulting plasmids were transformed into an mga-deleted GAS reporter strain (KSM148.174) containing a single-copy transcriptional fusion of gusA to the Mga-regulated Pemm, allowing direct quantitation of Mga-regulated activity based on a colorimetric assay (see Materials and Methods).
FIG. 1.
Random PCR mutagenesis screen used to identify domains involved in Mga-dependent transcriptional activation. Random mutations were generated in mga using the GeneMorph PCR mutagenesis kit (Stratagene). Mutated M6 mga alleles were placed on a plasmid under the constitutive promoter Pspac in an mga-deleted strain containing a promoter fusion of the Mga-regulated Pemm to a promoterless gusA reporter gene in the chromosome of M6 GAS. Resulting strains were plated onto THY plates containing X-Gluc to reveal clones deficient in transcriptional activation (light blue or white). Whole-cell lysates from clones showing a defect in activation were extracted, and Mga protein levels were determined using Western blot analysis. Plasmid DNA from clones producing wild-type levels of protein was isolated, sequenced, and aligned to the wild-type M6 mga gene to identify mutations.
Transformed GAS strains were plated onto media containing X-Gluc, which allowed visual determination of a defective Mga based on colony color (white or light blue) compared to blue seen with the wild-type control. Whole-cell lysates were prepared from clones chosen for further analysis and the steady-state level of Mga was determined using Western blot analysis. Finally, plasmid DNA from defective clones producing wild-type levels of protein was sequenced, and mutation sites were recorded (Table 3). Although identical mga mutations were found, possibly as a result of the amplification steps in E. coli, they were not included as separate entries in the overall results.
TABLE 3.
Mutants found from the random PCR mutagenesis screen
| Domain | Mutation | Phenotypea |
|---|---|---|
| DNA binding domain HTH-3 | F55C | LB |
| I62N | LB | |
| I71F | LB | |
| DNA-binding domain HTH-4 | S117T | W |
| S117N | W | |
| S119L | W | |
| T120K | W | |
| R123H | W | |
| Other | Q11R | LB |
| W12R/A38T | LB | |
| K33T | LB | |
| V30I/T139I | LB |
LB, light blue; W, white.
In total, 12 independent mutants were identified within the N-terminal mga fragment that resulted in either one or two amino acid changes per molecule and yet still produced wild-type levels of protein (Table 3). Eight mutants were identified that contained mutations within the known DNA binding domains HTH-3 and HTH-4 (16), resulting in colonies that were either light blue (HTH-3 mutations) or white (HTH-4 mutations). These activities correlated to the effects previously observed for mutations in the two Mga HTH domains (16) and served as a strong validation of the screen. Four additional mutations located outside of the known DNA binding regions were also identified that exhibited a light blue colony phenotype in the screen. Two pairs of mutants (Q11R, W12R/A38T and K33T, V30I/T139I) involved neighboring residues, suggesting that each might indicate potential functional domains in the N terminus of Mga.
Interestingly, no light blue or white colonies producing wild-type levels of protein were obtained from repeated screens of the C-terminal fragment of mga. In fact, 272 C-terminal mutants exhibiting a defect in Mga activity were analyzed via Western analysis and found to produce little to no detectable Mga (data not shown). Further analysis of a subset of these mutants found they each contained multiple amino acid changes, regardless of attempts to decrease the mutation frequency. Therefore, the C terminus of Mga appears to be quite sensitive to mutagenesis and, as a result, was not amenable to the screen used in this study.
Two mutations reside within a region conserved among Mga homologues.
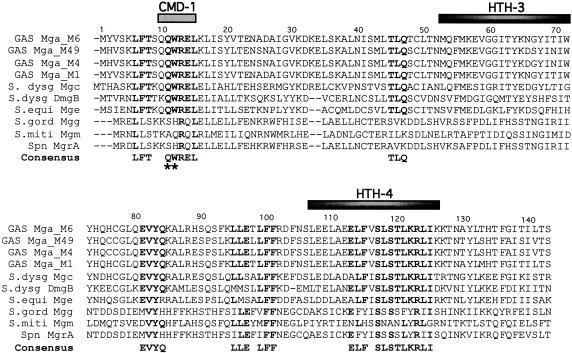
Conserved regions in protein families often represent important functional domains that are retained during evolution. Therefore, a protein sequence alignment was performed on Mga homologues from various streptococcal species, including Mga proteins representing divergent mga-1 and mga-2 alleles in the GAS, Mgc from S. dysgalactiae subsp. equisimilis, DmgB from S. dysgalactiae subsp. dysgalactiae, and MgrA from S. pneumoniae, as well as putative Mga-like transcriptional regulators found in the unfinished genomes of S. equi (Mge), S. gordonii (Mgg), and S. mitis (Mgm) via BLAST analysis (Fig. 2; see Materials and Methods).
FIG. 2.
Sequence alignment of Mga orthologues reveals conserved domains. A sequence alignment of the Mga orthologues from various streptococcal species, including GAS (serotypes M6, M49, M4, and M1), S. dysgalactiae (subsp. equisimilis and subsp. dysgalactiae), S. equi, S. gordonii, S. mitis, and S. pneumoniae, was used to derive a consensus sequence for conserved domains located within the first 143 amino acids of the proteins (see Materials and Methods). A conserved domain was defined as an area containing more than two consecutive residues exhibiting ≥70% identity among homologues. Amino acids identical to the consensus are in bold type. Black bars depict the locations of the two known DNA-binding domains in the GAS M6 Mga, while the gray bar denotes CMD-1. The two mutations found outside of the DNA-binding domains during the random mutagenesis screen are indicated (*).
A large region of conservation encompassed the essential HTH-4 domain of Mga, suggesting that this DNA-binding domain is likely serving a similar function in many of the orthologues (Fig. 2). Several other groups of conserved residues were also found by the alignment outside of the HTH domains (Fig. 2). Two of the four mutants identified in our activity screen (Table 3; K33T and V30I/T139I) did not fall within one of these conserved domains and were not investigated further in this study. However, the remaining two Mga mutants (Table 3; Q11R and W12R/A38T) contained mutations within a region of amino acid conservation (noted as asterisks in Fig. 2) spanning from the glutamine at position 11 to the leucine at position 15. The glutamine at position 10 also showed a high degree of conservation, and thus the region encompassing residues 10 to 15 was named the conserved Mga domain 1 (CMD-1).
CMD-1 of an M6 Mga (mga-1) is involved in transcriptional activation.
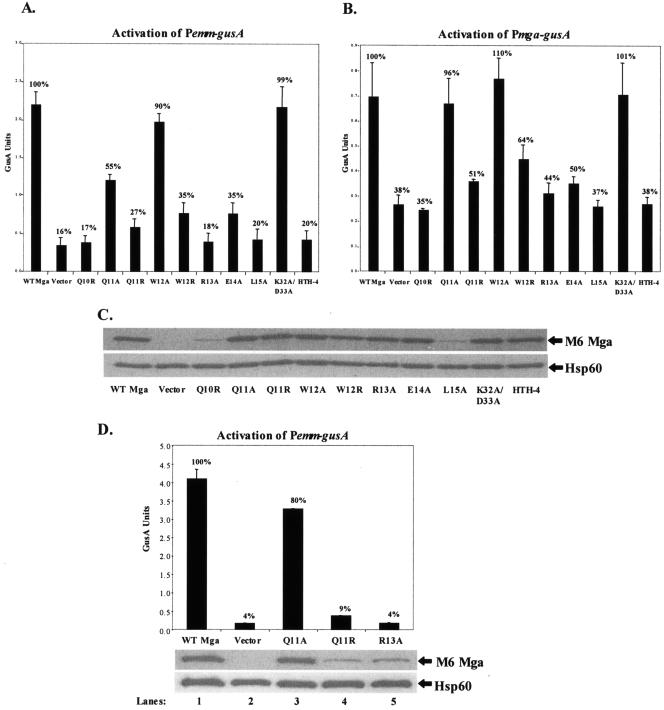
To establish whether the entire CMD-1 was important for Mga activity, we utilized site-specific mutagenesis to construct mutant mga alleles that encoded a single-amino-acid change in Mga at residues 10 to 15 (Q10R, Q11A, W12A, R13A, E14A, and L15A). In addition, the original Q11R mutant and a site-specific W12R mutant, which recapitulated the original W12R/A38T double mutant, were used. As in the initial screen, the resulting CMD-1 site-directed mutant alleles were cloned under the constitutive Pspac in pKSM318 to produce pQ10R, pQ11A, pQ11R, pW12A, pW12R, pR13A, pE14A, and pL15A (Table 2). A double Mga mutant, K31A/D32A (pKSM318.1H), which has been shown not to affect Mga-dependent activation, and an established HTH-4 mutation (pKSM318.4H), which leads to a DNA binding deficiency and loss of Mga-dependent activation, were constructed as controls (16). All of the plasmids were introduced into two mga-deleted serotype M6 GAS GusA reporter strains. The first, the Δmga Pemm-gusA strain KSM148.174 (27), determined Mga-regulated activity at a downstream promoter; while the second, the Δmga Pmga-gusA strain KSM231.310, assessed the ability to autoregulate.
Mga mutants Q10R, Q11A, Q11R, W12R, R13A, E14A, and L15A and the HTH-4 control mutant showed a reduction in activity at the mga-regulated promoter Pemm compared to wild-type Mga (Fig. 3A). The same mutants also showed a reduction in autoactivation at the native Pmga with the exception of Q11A, which appeared to have wild-type activity only in the Pmga strain (Fig. 3B). In contrast, mutation W12A and the negative control mutant K31A/D32A demonstrated wild-type promoter activity at both Pemm and Pmga (Fig. 3B). The steady-state levels of Mga detected in the strains expressing CMD-1 mutants in residues 11 to 14, as well as the control strains, were equivalent to the wild type compared to the loading control Hsp60 (Fig. 3C). However, flanking mutations at either end of CMD-1 (residues 10 and 15) show a reduction in protein level. Taken together, mutations in residues 11 through 14 in CMD-1 appear to affect activation without changing the overall steady-state levels of Mga, whereas amino acids 10 and 15 likely play a role in protein stabilization.
FIG. 3.
In vivo transcriptional activity of mutant M6 mga alleles. A) GusA activity of whole-cell lysates. Production of β-glucuronidase activity was determined for lysates from an mga-deleted Pemm-gusA reporter strain KSM148.174 or B) an mga-deleted Pmga-gusA reporter strain KSM231.310 containing plasmids expressing the following M6 Mga alleles from a constitutive Pspac promoter: wild-type M6 Mga, vector only, the M6 Mga mutants Q10R, Q11A, Q11R, W12A, W12R, R13A, E14A, and L15A. An arbitrary mutant K32A/D33A (no defect) and a known DNA-binding mutant in HTH-4 (defective) are included as controls. GusA units represent a measure of absorbance (A420)/protein concentration (μg/ml) and are the average of three independent experiments. C) Western analysis was performed on whole-cell lysates from the above samples using both an anti-Mga antibody for detection of protein levels (top) and an anti-Hsp60 antibody as a control for loading (bottom). D) Activity of mutants expressed from the native promoter. Activity levels for both wild-type and the M6 Mga mutants Q11A, Q11R, and R13A were examined for each allele when expressed from the native mga promoter using the Δmga Pemm-gusA reporter strain (top). Protein production was also determined via Western analysis (bottom) as described above. Percentages indicate level of GusA activity compared to that of the wild-type control.
As an independent verification of the role of CMD-1 in autoregulation at Pmga, the Q11A, Q11R, and R13A mutants in M6 Mga were produced from the native Pmga in the Δmga Pemm-gusA reporter strain KSM148.174 (Fig. 3D). As predicted by the Pmga reporter studies (Fig. 3B), those mutants showing a defect in Pmga-gusA activation (Q11R and R13A) did not produce wild-type levels of mutant protein from Pmga, whereas Q11A was normal for Pmga activation and showed levels of protein comparable to that of the wild-type control (Fig. 3D). Therefore, mutants in CMD-1 in M6 Mga are defective for both activation and autoregulation.
CMD-1 and HTH-4 are important for transcriptional activation in a divergent Mga.
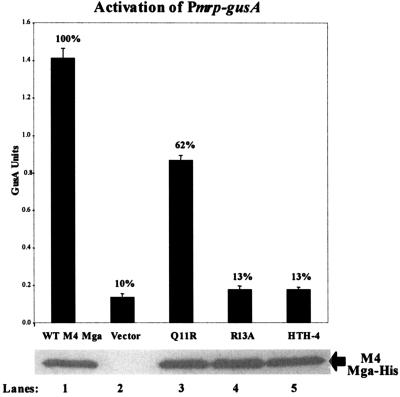
Since an mga-2 allele has been shown to functionally complement an mga-1 allele in vivo (2), we predicted that CMD-1 would play a similar role in a divergent Mga. To investigate the effects of CMD-1 mutations in a divergent Mga, we introduced mutations into the mga-2 allele from the serotype M4 strain AP4. Given that an antibody against the M4 Mga is not available, all of the alleles were modified to produce a carboxy-terminal 6X-His fusion to allow detection with anti-His monoclonal antibodies. Plasmids containing either the wild-type M4 mga-his allele or mga-his possessing the Q11R, R13A and HTH-4 mutations under their native Pmga4 promoter were transformed into the Pmrp-gusA GAS reporter strain KSM149. This strain has been used previously to study transcriptional activation by divergent Mga proteins (27).
GusA analysis demonstrated that the HTH-4 DNA-binding domain, which shows 100% identity within all Mga proteins from the GAS, is necessary for full transcriptional activation of Pmrp by a divergent Mga. Furthermore, a decrease in activation at Pmrp was also observed when the M4 Mga was mutated at either amino acid 11 or 13, implying that CMD-1 is important for Mga-specific activation in a divergent Mga as well (Fig. 4). Interestingly, all of the mutant alleles produced steady-state levels of Mga-His from Pmga4 equivalent to wild-type protein levels (Fig. 4), suggesting that the R13A and HTH-4 mutations are not defective for autoregulation as observed for the divergent M6 Mga (Fig. 3B).
FIG. 4.
In vivo transcriptional activity of mutant M4 mga alleles expressed from the native Pmrp promoter: GusA activity of whole-cell lysates (top). Production of β-glucoronidase activity was determined for lysates from an mga-deleted Pmrp-gusA reporter strain, KSM149, containing plasmids expressing the following different M4 mga alleles from the native Pmga promoter: wild-type M4 Mga (lane 1), vector only (lane 2), the M4 Mga mutants Q11R (lane 3) and R13A (lane 4), and a DNA-binding mutant in HTH-4 (lane 5). GusA units are a measure of the absorbance (A420)/protein concentration (μg/ml) and are the average of three independent experiments. Western analysis was performed on whole-cell lysates of each sample using an anti-His antibody for detection of Mga-His protein levels (bottom). Percentages indicate level of GusA activity compared to that of the wild-type control.
CMD-1 and HTH-4 are important for transcriptional activation in the Mga orthologue DmgB.
Since the CMD-1 and HTH-4 domains are important in both mga alleles from the GAS, we investigated whether these domains were also necessary for transcriptional activation in an Mga orthologue from another pathogenic streptococcus. A plasmid containing both the Mga-like regulator gene dmgB and the linked DmgB-regulated gene demB from S. dysgalactiae subsp. dysgalactiae (28) under the native PdmgB promoter were introduced into the Δmga M6 GAS strain JRS519 (17). Northern analysis was then used to monitor DmgB-mediated activation of demB in the Δmga GAS background (Fig. 5).
FIG. 5.
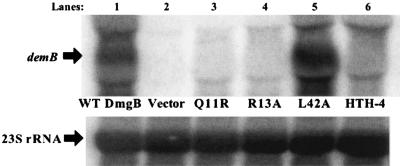
Transcriptional activation of demB by mutant Mga orthologue DmgB from S. dysgalactiae subsp. dysgalactiae. Northern analysis of transcript levels of the DmgB-regulated gene demB was determined using total RNA (5 μg) isolated from the GAS mga-inactivated M6 strain JRS519 harboring plasmids containing demB and the following dmgB alleles under the native dmgB promoter: wild-type dmgB (lane 1), vector only (lane 2), and the dmgB mutants Q11R (lane 3), R13A (lane 4), an arbitrary mutant L42A (lane 5), and an HTH-4 DNA-binding domain mutant (lane 6). Blots were stripped and reprobed with 23S rRNA to serve as a loading control (directly below). The blot shown is representative of data from three independent experiments.
Expression of wild-type dmgB leads to high levels of demB transcripts compared to either the vector alone (Fig. 5) or a deletion of dmgB from the plasmid (data not shown). Introduction of the HTH-4 mutation or the CMD-1 mutations Q11R and R13A into DmgB resulted in a dramatic reduction in demB transcript levels compared to the wild-type allele (Fig. 5). As a control, mutation of an arbitrary amino-terminal residue (L42A) of DmgB had little effect on its ability to regulate demB expression (Fig. 5). Overall, both CMD-1 and HTH-4 are necessary for activation of corresponding virulence genes not only in Mga, but also in other members of the Mga family of virulence regulators.
DNA-binding activity of CMD-1 Mga mutants.
Binding of Mga to promoter targets is essential for transcriptional activation of Mga-regulated genes in the GAS (16). To elucidate the contribution of CMD-1 to DNA binding, electrophoretic mobility shift assays were performed to determine the ability of Mga mutants to bind to Mga-regulated DNA targets in vitro. Plasmids containing either wild-type M6 mga-his (pKSM170) or the mutant M6 mga-his alleles Q11R (pKSM344), W12R (pKSM354), R13A (pKSM345), and E14A (pKSM355) were transformed into E. coli, and Mga proteins purified from each lysate. Increasing amounts of the purified Mga-His proteins were incubated with a constant amount of radiolabeled promoter probe corresponding to the M6 Mga-regulated promoters Pemm (Fig. 6A) and Pmga (Fig. 6B).
FIG. 6.
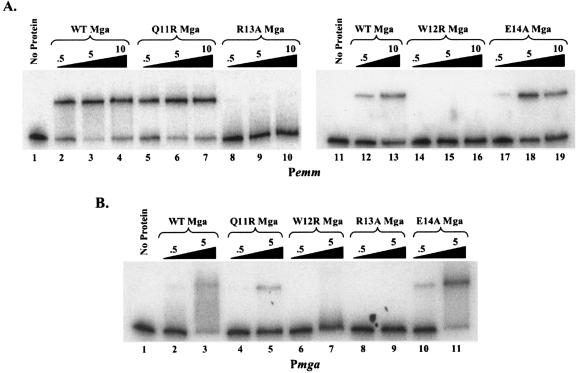
Electrophoretic mobility shift assays of CMD-1 Mga mutants binding to Mga-regulated promoters. (A) Electrophoretic mobility shift assays of Mga-regulated promoter Pemm. C-terminal 6X-His fusion proteins (Mga-His) were purified from E. coli lysates. Identical amounts of the radiolabeled promoter probe Pemm were incubated for 15 min at 16°C with an increasing amount (0.5, 5, and 10 μg) of either the wild-type (lanes 2 to 4 and 12 to 13), Q11R (lanes 5 to 7), R13A (lanes 8 to 10), W12R (lanes 14 to 16), E14A (lanes 17 to 19), or no M6 Mga-His (lanes 1 and 11) prior to separation on a 5% polyacrylamide gel. (B) Electrophoretic mobility shift assays of the native Pmga promoter were performed as described above using 0.5 and 5 μg of the wild type (lanes 2 and 3), Q11R (lanes 4 and 5), W12R (lanes 6 and 7), R13A (lanes 8 and 9), E14A (lanes 10 and 11), or no Mga-His protein (lane 1).
EMSA reactions containing the wild-type Mga (Fig. 6A, lanes 2 to 4, 12, and 13, and Fig. 6B, lanes 2 to 3) and mutant Mga proteins Q11R (Fig. 6A, lanes 5 to 7, and Fig. 6B, lanes 4 to 5) and E14A (Fig. 6A, lanes 17 to 19, and Fig. 6B, lanes 10 to 11) showed reduced mobility of both promoter probes. However, neither W12R (Fig. 6A, lanes 14 to 16, and Fig. 6B lanes, 6 to 7) nor R13A (Fig. 6A, lanes 8 to 10, and Fig. 6B, lanes 8 to 9) were able to bind at wild-type levels to either of the two probes. Thus, two of the CMD-1 mutants (W12R and R13A) were defective for binding to DNA targets, whereas two other flanking CMD-1 mutants (Q11R and E14) exhibited normal DNA-binding activity. Therefore, a loss of DNA binding alone does not explain the defect in transcriptional activation observed with some CMD-1 mutations.
DISCUSSION
HTH-4 and CMD-1 are functional domains of Mga.
Despite its established role in the pathogenesis of GAS infections, we currently know very little about how Mga functions. Previous studies using a serotype M6 Mga (mga-1) identified a minor (HTH-3) and a major (HTH-4) helix-turn-helix DNA-binding domain within the amino terminus of Mga (16). Importantly, mutations in HTH-4 led to a defect in M6 Mga-dependent activation in vivo and established DNA binding as an essential function of Mga. Finding additional functional domains in Mga would provide insights into its mechanism of action.
To address this issue, we utilized a genetic screen to look for random mutations in the serotype M6 Mga (mga-1) that lead to a defect in its ability to activate Mga-regulated gene transcription in vivo. The screen identified both the HTH-3 and HTH-4 DNA-binding domains of M6 Mga (Fig. 2), providing further evidence of their importance and acting as a strong validation of the screen. The analysis also revealed two amino acids (Q11 and W12) involved in M6 Mga activity that were conserved among divergent Mga proteins in the GAS and orthologues found in other pathogenic streptococci. Site-directed mutagenesis within the conserved Mga domain 1 (CMD-1; amino acids 10 to 15) verified the importance of CMD-1 for transcriptional activation of M6 Mga-regulated genes (emm) as well as its own autoregulation (mga) in this background (Fig. 3). Therefore, CMD-1 represents a new Mga functional domain involved in its ability to regulate virulence gene expression in the GAS.
To date, most of the functional studies performed on Mga have been done using the serotype M6 (mga-1) allele (16, 18). Thus, it was important to see if functional domains identified in the M6 Mga were also required in a divergent M4 Mga (mga-2). Our recent studies have found that some amino acids important for M4 Mga (mga-2) activity did not serve the same function in a divergent M6 Mga (mga-1) (27). However, site-directed mutations in both the HTH-4 and CMD-1 domains of the M4 Mga (mga-2) resulted in a protein that was unable to activate transcription of the M4 Mga-regulated gene mrp (Fig. 4). Therefore, both motifs appear to serve similar roles in divergent mga alleles and represent conserved functional domains in all Mga proteins.
Functional role for CMD-1 in Mga activity.
Given that CMD-1 is important for Mga-dependent transcriptional activation, exactly how the domain contributes to Mga function is of particular interest. The mutational analysis provided some clues as to the role of specific residues. It was clear from the mutagenesis of CMD-1 that mutations in either of the two amino acids (position 10 and 15) flanking the conserved region showed diminished activation due to low steady-state protein levels (Fig. 3C), suggesting that these two residues are likely important in overall Mga stability. Most alanine substitutions within the rest of CMD-1 in M6 Mga resulted in a significant decrease in activity by as much as 83% for Pemm and 65% for Pmga without affecting the levels of protein (Fig. 3AB). Similar trends were seen for those CMD-1 mutations tested in the divergent M4 Mga, although differences in the degree of reduction were observed for the Q11R mutation (Fig. 4). One M6 Mga mutant (W12A) did not show a decrease in transcription at either promoter; however, when W12 was mutated to an arginine instead of an alanine, a loss of activity was observed. This implies that, at least for this residue in CMD-1, the ability of Mga to activate transcription is contingent upon the particular amino acid at that position, possibly reflecting a charged or polar interaction.
Because DNA binding is essential for Mga activity (16), CMD-1 M6 Mga mutants were tested for their ability to bind probes corresponding to the Mga-regulated Pemm and Pmga promoters. It was expected that all CMD-1 mutations would be either wild type or defective in their ability to bind to regulated promoter targets. Surprisingly, two of the four transcriptionally defective mutants tested (Q11R and E14A) retained the ability to bind DNA, while the other two mutants (W12R and R13A) did not (Fig. 6A and B). The inability of the W12R mutant to bind normally may be dependent upon the amino acid change at that position as discussed above. Since a W12A mutation at this position shows wild-type Mga activity (Fig. 3A and B), it is predicted to retain normal binding activity as well. The other binding mutant, R13A, occurs at an arginine residue that is 100% conserved among Mga orthologues (Fig. 2), and we can only speculate what effects different mutations at R13 would have on the ability to bind or activate Mga-regulated promoters. Therefore, at least two residues in CMD-1 are important for DNA binding in addition to the established HTH-3 and HTH-4 domains.
The ability of two mutants (Q11R and E14A) to bind DNA normally while still being defective in transcriptional activation in vivo clearly suggests that portions of CMD-1 contribute to Mga activity independently of DNA binding. Secondary-structure predictions (Jpred; http://www.compbio.dundee.ac.uk/∼www-jpred) suggest that CMD-1 is part of an alpha-helix. Mutants at positions 11 and 14 not only demonstrate a similar phenotype, but also would reside on the same face of the helix. This aspect of CMD-1 potentially represents a novel function for this regulator, and further study may provide us with new insights into the mechanism of Mga regulation.
Autoregulation and divergent Mga proteins.
The M6 Mga (mga-1) has been shown to bind directly to its own promoter, resulting in activation of mga expression and amplification of the Mga response (7, 16, 18). In this study, constitutive expression of M6 Mga CMD-1 and HTH-4 mutants unable to activate Pemm also demonstrated a corresponding defect in autoactivating Pmga (Fig. 3AB). Furthermore, expression of several CMD-1 M6 Mga mutants from their native M6 Pmga did not produce wild-type levels of protein (Fig. 3D), further supporting a direct role of an active Mga in its own regulation.
Interestingly, the loss of Mga activity caused by the same CMD-1 mutations as well as the HTH-4 mutation in the divergent M4 Mga (mga-2) had no effect on their autoregulated expression from their native M4 Pmga (Fig. 4). In fact, we observed a similar lack of autoregulation in a recent study of naturally occurring mutants introduced into M4 Mga expressed from the M4 Pmga (27). Therefore, it appears that normal expression of M4 Mga from its own promoter is not dependent on an active Mga. Promoter analysis of Pmga from an M49 (mga-2) strain found significant differences at the nucleotide level compared to the same region from an M6 (mga-1) strain (21), including a nine-nucleotide insert into Mga binding site 2 (16). Previous studies suggested autoregulation in the M49 system based on Northern analysis (21). Given that the M4 and M49 strains share 99.5% sequence identity across Pmga, it will be interesting to determine whether all mga-2 alleles share a common mechanism of mga regulation.
Another interesting observation involved the M6 Mga mutant Q11A produced from a constitutive promoter, which was transcriptionally defective only at Pemm and not Pmga (Fig. 3A and B), suggesting that it is possible to unlink the ability of an M6 Mga to activate downstream promoters and autoactivation. This hypothesis is supported by the differences in the number, size, and location of Mga binding sites at each promoter compared to the start of transcription (14, 16, 18).
Conserved functional domains define an Mga family of virulence regulators.
Regions of amino acid conservation found between related proteins will often highlight those areas that are indispensable for function in the cell. In this study, we propose a new family of transcriptional regulators found within various pathogenic streptococcal species, including established virulence regulators from S. dysgalactiae and S. pneumoniae, based on their sequence homology to Mga proteins from the GAS. Even though individual members can vary considerably from one to another, regions of 70 to 100% identity were observed in the different regulatory proteins. Two such conserved domains, CMD-1 and HTH-4, were subsequently found in our genetic screen as being essential for transcriptional activation in all Mga alleles tested. Based on these results, one would predict that these regions might likely serve similar roles in other members of this family. Using the Mga-like transcriptional regulator DmgB from S. dysgalactiae subsp. dysgalactiae to test this hypothesis, we found that mutations in both the conserved HTH-4 and CMD-1 domains resulted in an inactive DmgB, as predicted from our results with Mga (Fig. 5).
Due to technical reasons, our screen only investigated the amino-terminal 150 residues of Mga. Thus, there may be a number of domains in the full-length molecules that will provide interesting targets for further analysis. Although functional differences are bound to exist among the proteins to correspond with the variations in regulated genes, vast amounts of knowledge applicable to the entire Mga family can be gained through exploration of the conserved regions within family members.
Acknowledgments
We thank Bengt Guss for kindly providing the S. dysgalactiae dmgB demB clones. Critical reading by Deborah Ribardo, Audry Almengor, and Temekka Leday as well as assistance with graphic illustration by Richard Bonnette is greatly appreciated.
This work was supported by NIH/NIAID Public Health Service award R01-AI47928 to K.S.M.
REFERENCES
- 1.Almengor, A. C., and K. S. McIver. 2004. Transcriptional activation of sclA by Mga requires a distal binding site in Streptococcus pyogenes. J. Bacteriol. 186:7847-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, G., K. McIver, L. O. Heden, and J. R. Scott. 1996. Complementation of divergent mga genes in group A streptococcus. Gene 175:77-81. [DOI] [PubMed] [Google Scholar]
- 3.Bessen, D. E., A. Manoharan, F. Luo, J. E. Wertz, and D. A. Robinson. 2005. Evolution of transcription regulatory genes is linked to niche specialization in the bacterial pathogen Streptococcus pyogenes. J. Bacteriol. 187:4163-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Facklam, R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geist, R. T., N. Okada, and M. G. Caparon. 1993. Analysis of Streptococcus pyogenes promoters by using novel Tn916-based shuttle vectors for the construction of transcriptional fusions to chloramphenicol acetyltransferase. J. Bacteriol. 175:7561-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geyer, A., and K. H. Schmidt. 2000. Genetic organisation of the M protein region in human isolates of group C and G streptococci: two types of multigene regulator-like (mgrC) regions. Mol. Gen. Genet. 262:965-976. [DOI] [PubMed] [Google Scholar]
- 9.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 10.Hemsley, C., E. Joyce, D. L. Hava, A. Kawale, and A. Camilli. 2003. MgrA, an orthologue of Mga, acts as a transcriptional repressor of the genes within the rlrA pathogenicity islet in Streptococcus pneumoniae. J. Bacteriol. 185:6640-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollingshead, S. K., J. Arnold, T. L. Readdy, and D. E. Bessen. 1994. Molecular evolution of a multigene family in group A streptococci. Mol. Biol. Evol. 11:208-219. [DOI] [PubMed] [Google Scholar]
- 12.Hollingshead, S. K., T. L. Readdy, D. L. Yung, and D. E. Bessen. 1993. Structural heterogeneity of the emm gene cluster in group A streptococci. Mol. Microbiol. 8:707-717. [DOI] [PubMed] [Google Scholar]
- 13.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 14.McIver, K. S., A. S. Heath, B. D. Green, and J. R. Scott. 1995. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J. Bacteriol. 177:6619-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIver, K. S., A. S. Heath, and J. R. Scott. 1995. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 63:4540-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIver, K. S., and R. L. Myles. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol. Microbiol. 43:1591-1602. [DOI] [PubMed] [Google Scholar]
- 17.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McIver, K. S., A. S. Thurman, and J. R. Scott. 1999. Regulation of mga transcription in the group A streptococcus: specific binding of Mga within its own promoter and evidence for a negative regulator. J. Bacteriol. 181:5373-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada, N., R. T. Geist, and M. G. Caparon. 1993. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol. Microbiol. 7:893-903. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Podbielski, A., A. Flosdorff, and J. Weber-Heynemann. 1995. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect. Immun. 63:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Podbielski, A., J. A. Peterson, and P. Cleary. 1992. Surface protein-CAT reporter fusions demonstrate differential gene expression in the vir regulon of Streptococcus pyogenes. Mol. Microbiol. 6:2253-2265. [DOI] [PubMed] [Google Scholar]
- 23.Ribardo, D. A., and K. S. McIver. 2003. amrA encodes a putative membrane protein necessary for maximal exponential phase expression of the Mga virulence regulon in Streptococcus pyogenes. Mol. Microbiol. 50:673-685. [DOI] [PubMed] [Google Scholar]
- 24.Scott, J. R., P. C. Guenthner, L. M. Malone, and V. A. Fischetti. 1986. Conversion of an M− group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J. Exp. Med. 164:1641-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stenberg, L., P. O'Toole, and G. Lindahl. 1992. Many group A streptococcal strains express two different immunoglobulin-binding proteins, encoded by closely linked genes: characterization of the proteins expressed by four strains of different M-type. Mol. Microbiol. 6:1185-1194. [DOI] [PubMed] [Google Scholar]
- 26.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 27.Vahling, C. M., and K. S. McIver. 2005. Identification of residues responsible for the defective virulence gene regulator Mga produced by a natural mutant of Streptococcus pyogenes. J. Bacteriol. 187:5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasi, J., L. Frykberg, L. E. Carlsson, M. Lindberg, and B. Guss. 2000. M-like proteins of Streptococcus dysgalactiae. Infect. Immun. 68:294-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whatmore, A. M., V. Kapur, D. J. Sullivan, J. M. Musser, and M. A. Kehoe. 1994. Noncongruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol. Microbiol. 14:619-631. [DOI] [PubMed] [Google Scholar]