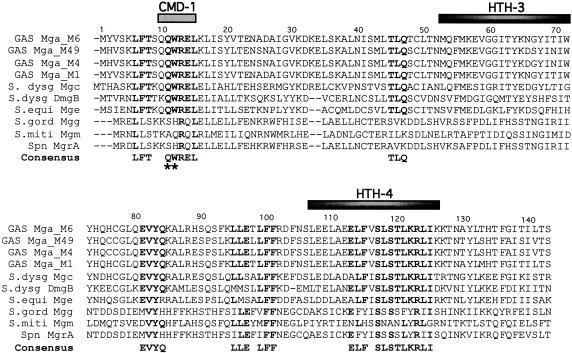
FIG. 2.
Sequence alignment of Mga orthologues reveals conserved domains. A sequence alignment of the Mga orthologues from various streptococcal species, including GAS (serotypes M6, M49, M4, and M1), S. dysgalactiae (subsp. equisimilis and subsp. dysgalactiae), S. equi, S. gordonii, S. mitis, and S. pneumoniae, was used to derive a consensus sequence for conserved domains located within the first 143 amino acids of the proteins (see Materials and Methods). A conserved domain was defined as an area containing more than two consecutive residues exhibiting ≥70% identity among homologues. Amino acids identical to the consensus are in bold type. Black bars depict the locations of the two known DNA-binding domains in the GAS M6 Mga, while the gray bar denotes CMD-1. The two mutations found outside of the DNA-binding domains during the random mutagenesis screen are indicated (*).