Abstract
Meiotic recombination occurs preferentially at certain regions in the genome referred to as hot spots. The number of hot spots known in humans has increased manifold in recent years. The identification of these hot spots in humans is of great interest to population and medical geneticists since they influence the structure of Linkage Disequilibrium and Haplotype blocks in human populations, whose patterns have applications in mapping disease genes. HUMHOT is a web-based database of Human Meiotic Recombination Hot Spots. The database comprises DNA sequences corresponding to the hot spot regions from the literature that have been mapped to a high resolution (<4 kb) in humans. It also provides flanking sequence information for the hot spot region along with references describing the hot spot. The database can be queried based on hot spot identity, chromosome position or by homology to user-defined sequences. It is also updated with new hot spot sequences as they are discovered and provides hyperlinks to commonly used tools for estimating recombination rates, performing genetic analysis and new advances in our understanding of meiotic hot spots. Public access to the HUMHOT database is available at http://www.jncasr.ac.in/humhot.
INTRODUCTION
Meiotic recombination is initiated at the prophase stage of meiosis I through the formation of double strand breaks (DSBs) by the Spo11 endonuclease (1,2). The non-random distribution of DSBs that initiate the recombination events results in the formation of hot spots where the recombination frequency between markers exceeds the average recombination frequency for the entire genome. A segment of DNA that undergoes recombination at the genome average rate can also be considered a hot spot if it is embedded in a recombinationally suppressed region of the genome. Hot spots have been observed to be 1–3 kb wide in yeast, mice and humans (3) and unlike the chi sequence in Escherichia coli (4), no primary DNA sequence determinant of hot spot activity has been identified in eukaryotes. In recent years, a large number of human meiotic recombination hot spots have been identified due to the development of methods to analyze sperm DNA (5) and population genetics approaches (6). Comparison of the molecular features of meiotic recombination hot spots between yeast, mice and humans reveals a high degree of similarity and suggests overlapping roles for both the DNA sequence and chromatin configuration in the establishment of a hot spot (3,7,8). It has been estimated that the human genome is likely to contain as many as 50 000 meiotic hot spots on the basis of the distribution of hot spots in the MHC region and from the large-scale identification of recombination hot spots on specific chromosomes (3).
Recombination hot spots in humans frequently delimit the extent of haplotype blocks in populations as observed from the correlation between sperm crossover hot spots and regions showing LD breakdown on two different chromosomal locus (9–11). This has resulted in a role for meiotic hot spots in association mapping of loci contributing to phenotypic traits since they help define haplotype blocks that reduce the number of markers required for such analysis. However, a few exceptions in this paradigm have also emerged recently (9,12) which also support the argument that hot spots are very fluid features of the genome on evolutionary time scales (13).
The HUMHOT database has been conceived with the idea to store all meiotic recombination hot spot sequences identified in humans by a database managing system. It currently stores 132 human meiotic recombination hot spot sequences identified till date through sperm typing or population genetics based approaches. Users can query the database based on hot spot identity (locus name) or chromosome number. It is also possible to perform a homology search, which determines whether the sequence submitted exists as a hot spot sequence in the database. The database stores various details for every hot spot sequence, such as locus name, chromosome number, hot spot sequence in FASTA format, flanking sequence information in GCG format, hyperlinks to the respective reference papers on PubMed, and accession numbers to get further sequence details from the GenBank or ENCODE databases, as the case may be. The sequences are also available in a downloadable flat file version so that users can easily store them and perform additional operations on them. The database also provides information on bibliographic knowledge about hot spots and recent published literature in this field.
DATABASE STRUCTURE
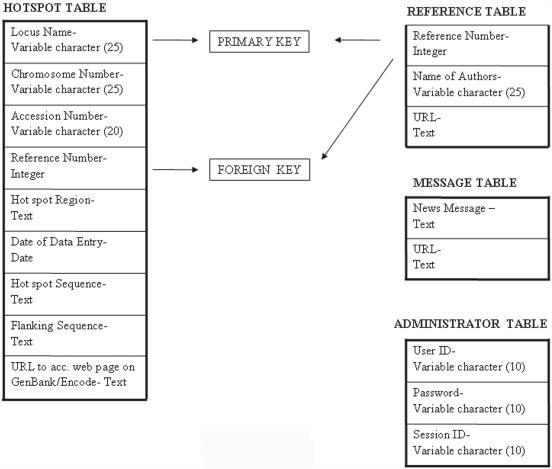
The Database Management System that we are using is PostgreSQL since it is freely available and also used by the server at JNCASR. The software has been coded with PHP, HTML and Javascript as the higher languages. The database houses information about the various hot spot sequences in the form of tables (Figure 1) and flat files. The table ‘Hotspot’ which houses details of the sequences has the following information stored for every such sequence: Locus Name, Chromosome Number, Accession Number, Hot spot Region in base pairs, Date of Data Entry, Reference ID, Hot spot Sequence and Flanking Sequence information and URL to the corresponding accession web page in the GenBank. Locus Name is the primary key of the above table. Another table called the ‘Reference Table’ stores the Reference ID, name of the author and the URL to the web page where the particular paper is available. The primary key of this table is the Reference ID, which is also the foreign key for the Hotspot table. The ‘Administrator’ table houses information about the Administrator and maintain session ids. The Message table stores the Recombination news messages and their respective URL's. There is provision for storing three news items at a time.
Figure 1.
Database design and operation. The database has been designed using PostgreSQL. It comprises four tables—Hotspot table which stores details of all the sequences, Reference table for storing reference articles describing the hot spots, Message table for inserting recombination news and the Administrator table which provides a secure login feature for the Administrator and maintains the session id. The database is quick in responding to a query since the queries to the database are being carried out simultaneously on a maximum of two tables (Hotspot table, Reference table) only.
DATABASE ACCESS AND WEB QUERY INTERFACE
The website is extremely user friendly and offers easy navigability options across the web pages through a drop down horizontal menu bar, with appropriate directional and error messages at each step.
Search features
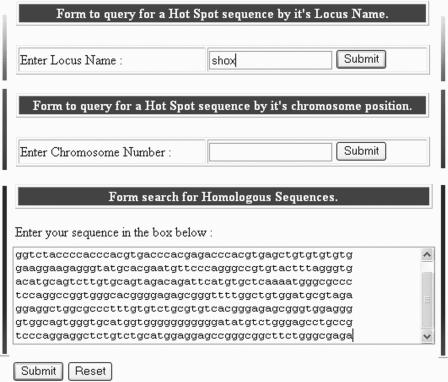
The HUMHOT web interface provides access to the database contents in three ways through the ‘search database’ option (Figure 2). The user can either enter the hot spot locus name or a chromosome number or use the homology interface that queries all sequences in the HUMHOT database. In cases in which a search identifies more than one database entry, the names of all corresponding hits are displayed for the user to choose (Figure 3). The locus name should match the hot spot name as entered in the database while the chromosome number entered can specify the chromosome arm also. For a given match, the hot spot sequence in FASTA format and the sequence flanking the hot spot in GCG format are presented along with links to the reference describing the hot spot and through the accession number to the relevant database (GenBank, ENCODE). A printable version and flat file format of the hot spot and flanking sequence can also be generated.
Figure 2.
Screen shot showing the multiple query forms generated from the drop down menu under the ‘search database’ option. Selecting a menu from the ‘Search database’ drop down box opens a simple form in which the user can specify the hot spot locus name, chromosome number or paste a query sequence to search the database.
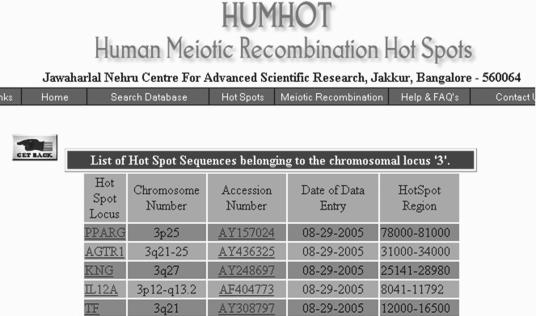
Figure 3.
Sample output of the HUMHOT database following search for hot spots on chromosome 3. The output lists all hot spots in the database on chromosome 3. Selecting any of the hot spots under the field hot spot locus opens up a new page containing the hot spot sequence, flanking sequence, reference and accession number. The hot spot region indicated is with respect to the flanking sequence.
Additional database contents
The website also provides information about various aspects of meiotic hot spots, such as different classes of hot spots, motifs associated with hot spots, molecular features of hot spots, methods used to map hot spots and an illustration of the meiotic recombination process in different species. The ‘Recombination Tool Box’ provides easy access to different software and websites on the internet dedicated to computing recombination rates and other kinds of genetic analysis. The ‘Useful Links’ button on the menu bar allows users to access other websites which could be helpful for DNA sequence analysis and sequence format conversions, since the database comprises DNA sequences. Users can also know as to when the database was last updated by clicking on the date, which appears right below the ‘Last Updated’ button on the home page. This action would open a new page where the details of all of the sequences that were updated would be displayed in a tabular form. To keep visitors updated on recent information on hot spots, the ‘Recombination News’ section has been provided. The scrolling news message is hyperlinked to the respective article on PubMed.
FUTURE DEVELOPMENTS
The website provides secure access to the Administrator to modify and update the database as and when new human meiotic hot spot sequences are identified. Researchers working on meiotic recombination are also welcome to submit published data on new human meiotic hot spot sequences by email to the corresponding authors. We plan to expand the HUMHOT database to include all human meiotic hot spot sequences along with an additional information on the methods used to map the hotspots. We also intend to improve the usefulness of the database by including a direct link from the hot spot regions to the haplotype map available for the corresponding chromosomal region on the human HapMap webpage. This would be done in the near feature.
CITING HUMHOT
Authors who make use of the HUMHOT database as a tool for their published research can cite this paper as reference, and quote the HUMHOT home page URL, http://www.jncasr.ac.in/humhot.
Acknowledgments
The authors thank Gilean McVean, University of Oxford for providing the hot spot positions in the chromosome 20 dataset and Xuegong Zhang, Tsinghua University for providing recombination rate estimates for genes in the Seattle SNP database. The authors are thankful to Ms Sheethal (Network Administrator, JNCASR, Bangalore) for assistance with the uploading and better working of website. Funding to pay the Open Access publication charges for this article was provided by JNCASR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sun H., Treco D., Schultes N.P., Szostak J.W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 2.Keeney S., Giroux C.N., Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 3.Kauppi L., Jeffreys A.J., Keeney S. Where the crossovers are: recombination distributions in mammals. Nature Rev. Genet. 2004;5:413–424. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- 4.Smith G.R., Amundsen S.K., Dabert P., Taylor A.F. The initiation and control of homologous recombination in E.coli. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1995;347:13–20. doi: 10.1098/rstb.1995.0003. [DOI] [PubMed] [Google Scholar]
- 5.Hubert R., MacDonald M., Gusella J., Arnheim N. High resolution localization of recombination hot spots using sperm typing. Nature Genet. 1994;7:420–424. doi: 10.1038/ng0794-420. [DOI] [PubMed] [Google Scholar]
- 6.Stumpf M.P., McVean G.A. Estimating recombination rates from population genetic data. Nature Rev. Genet. 2003;4:959–968. doi: 10.1038/nrg1227. [DOI] [PubMed] [Google Scholar]
- 7.Jeffreys A.J., Neumann R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nature Genet. 2002;31:267–271. doi: 10.1038/ng910. [DOI] [PubMed] [Google Scholar]
- 8.de Massy B., Rocco V., Nicolas A. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in S. cerevisiae. EMBO J. 1995;14:4589–4598. doi: 10.1002/j.1460-2075.1995.tb00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeffreys A.J., Neumann R., Panayi M., Myers S., Donnelly P. Human recombination hot spots hidden in regions of strong marker association. Nature Genet. 2005;37:601–606. doi: 10.1038/ng1565. [DOI] [PubMed] [Google Scholar]
- 10.Jeffreys A.J., Kauppi L., Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nature Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein D.B. Islands of linkage disequilibrium. Nature Genet. 2001;2:109–111. doi: 10.1038/ng1001-109. [DOI] [PubMed] [Google Scholar]
- 12.Kauppi L., Stumpf M.P., Jeffreys A.J. Localized breakdown in linkage disequilibrium does not always predict sperm crossover hot spots in the human MHC class II region. Genomics. 2005;86:13–24. doi: 10.1016/j.ygeno.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Clark A.G. Hot spots unglued. Nature Genet. 2005;37:563–564. doi: 10.1038/ng0605-563. [DOI] [PubMed] [Google Scholar]