Abstract
The Gene Ontology (GO) project (http://www.geneontology.org) develops and uses a set of structured, controlled vocabularies for community use in annotating genes, gene products and sequences (also see http://song.sourceforge.net/). The GO Consortium continues to improve to the vocabulary content, reflecting the impact of several novel mechanisms of incorporating community input. A growing number of model organism databases and genome annotation groups contribute annotation sets using GO terms to GO's public repository. Updates to the AmiGO browser have improved access to contributed genome annotations. As the GO project continues to grow, the use of the GO vocabularies is becoming more varied as well as more widespread. The GO project provides an ontological annotation system that enables biologists to infer knowledge from large amounts of data.
INTRODUCTION
The Gene Ontology (GO) project (http://www.geneontology.org) is a collaborative effort to construct and use ontologies to facilitate the biologically meaningful annotation of genes and their products in a wide variety of organisms. Groups participating in the project include the major model organism databases and other bioinformatics resource centers.
The GO Ontologies provide a systematic language, or ontology (1–4), for the description of attributes of genes and gene products, in three key domains that are shared by all organisms, namely molecular function, biological process and cellular component (5–10); sequence features are covered by the Sequence Ontology, maintained separately from the GO ontologies (11).
The GO annotations have proven to be remarkably useful for the mining of functional and biological significance from very large datasets, such as microarray results. The GO also facilitates the organization of data from novel, as well as fully annotated, genomes and the comparison of biological information between clade members and across clades.
IMPROVEMENTS IN GO CONTENT
From its inception, the GO project has developed its ontologies for the purpose of gene product annotation. To this end, the Gene Ontology is dynamic: existing terms and relationships are augmented, refined and reorganized as the current state of biological knowledge advances. Major improvements have been made over the past 2 years in several areas of the ontology, often in consultation with experts in relevant subject areas. The Plant-Associated Microbe Gene Ontology (PAMGO) Interest Group collaborated with the GO Consortium to produce a new set of terms representing pathogenic and symbiotic processes (also see below). With help from representatives of the BioCyc databases, the GO representation of metabolism was split into cellular and organismal processes. The cell cycle node was extensively reworked and is undergoing further improvement. Finally, high level terms were added to the cellular component ontology to better categorize terms representing the constituents of cells. A summary of the current ontology content is shown in Table 1.
Table 1.
Current status of GO
| Biological process terms | 9805 |
| Molecular function terms | 7076 |
| Cellular component terms | 1574 |
| Sequence Ontology terms | 963 |
| Genomes with annotationa | 30 |
| Annotated gene products | |
| Total | 1618 739 |
| Electronic only | 1460 632 |
| Manually curated | 158 107 |
aExcludes annotations from UniProt, which represent 261 annotated proteomes.
MANAGING CONTENT CHANGES
All changes to the ontologies are centrally coordinated by the GO Editorial Office (located at the European Bioinformatics Institute, Hinxton, UK). Changes are proposed by GO curators, model organism database annotators and other interested parties throughout the biological community. GO curators have adapted the online tracking system provided by SourceForge to document progress (see http://geneontology.sourceforge.net/); as of September 1, 2005, over 2800 items have been posted, of which over 2100 have led to changes in the ontologies.
The model organism database curators who use GO terms intensively for gene product annotation play a key role in guiding the development of GO. To complement their input, the GO Consortium strives to involve members of the biological research community in the ontology development process. Experts in various biomedical fields provide thorough, detailed knowledge of their particular topics that complements GO curators' understanding of existing GO structures and conventions.
To promote communication among these various contributors and ensure consistency within the ontology, the GO Consortium has established Curator Interest Groups and has initiated a series of meetings devoted to ontology content; both provide mechanisms to focus on areas within the ontologies that are likely to require extensive additions or revisions. Curator Interest Group membership is open not only to Consortium members, but also to community experts in the field. A list of the 29 current Interest Groups can be found at http://www.geneontology.org/GO.interests.shtml.
GO content meetings serve to bring GO curators and biologists together to resolve specific sub-trees of the GO structure. Many of the recent improvements in GO stem from the first content meeting, held in August 2004, where members of the GO group and domain experts in plant pathogens (PAMGO), the cell cycle and metabolism participated.
PAMGO: a case study
The successful interaction between the PAMGO group and GO curators provides a model that the GO Consortium will use to involve research communities to cover a number of additional topics in the future. The PAMGO Interest Group (http://pamgo.vbi.vt.edu/) was formed in 2004 to develop new higher level biological process terms for annotating gene products of various microbes (bacteria, oomycetes, fungi and nematodes) involved in pathogenic interactions with plants. Prior to the August 2004 GO content meeting, the PAMGO group drafted a set of high level terms to represent the range of host-microbe interactions, from mutualism to parasitism, for any microbial species and for animal as well as plant hosts. The proposal generated intensive discussion during and after the GO Content meeting, and three modified options were considered at a GO Consortium meeting in October 2004. A final ‘tree’ of terms, including 35 newly created terms, was resubmitted to GO in December 2004 and incorporated into the ontology structure in January 2005. The final set of terms is thus a synthesis of PAMGO's original submission and contributions from the GO Consortium, and the result of a process that included broad-ranging discussion across the wider GO community about the definitions of high level terms.
INCREASING ANNOTATION COVERAGE AND QUALITY CONTROL
Alongside the development of GO ontology content, the use of GO terms for gene product annotation has increased substantially. Annotation data are now subject to checks to maintain file format integrity and avoid redundancy, and GO Consortium member groups are developing measures to assess the accuracy and consistency of annotations made by different individuals or groups [for example see (12,13)].
Furthermore, the GO Consortium has recently begun an effort to actively support new groups seeking to use GO for gene product annotation and to make the resulting annotation data available to the public as part of the GO repository. GO annotations are now available for over 30 genomes [plus many additional species, including 261 proteomes, via UniProt (14)], with recent additions including chicken and several prokaryotes.
GO ANNOTATION CAMPS
This attention to annotation outreach has led the GO Consortium to initiate a series of meetings devoted to GO annotation practices. These meetings, known as ‘Annotation Camps’, review and refine the approaches that the GO Consortium now takes to improve the coverage, accuracy and precision of GO annotation data. At the first Annotation Camp, held in June 2004, GO Consortium members focused on developing and maintaining consistent annotation practices within and among groups. The second Annotation Camp, held in June 2005, was larger and open to non-members (about two-thirds of the participants), and thus served to help educate people unfamiliar with the GO system, as well as continuing to work toward the consistency goals of the first Camp. Each Annotation Camp introduced the basic organization of the GO and covered a number of practical aspects of its use. A key component of the Annotation Camps was the review of example papers by working groups, to improve the consistency of gene product annotation based on literature.
In addition to the Consortium-wide Annotation Camps, some GO Consortium members, such as The Institute for Genomic Research, run their own annotation courses and make annotation tools publicly available; individual database curators may also learn directly from ‘mentors’ with extensive experience using the GO system.
SOFTWARE DEVELOPMENTS
The GO Consortium provides software tools to navigate, use and manipulate the GO terms and annotations. Many new features have been added to the Java-based editing tool DAG-Edit (http://godatabase.org/dev/), and its successor, OBO-Edit, is in beta testing. OBO-Edit adds support for many of the advanced features of ontology languages, such as OWL. GO and OBO-Edit are also closely coordinated with the development of Obol, a formal language for specifying ontology terms (15).
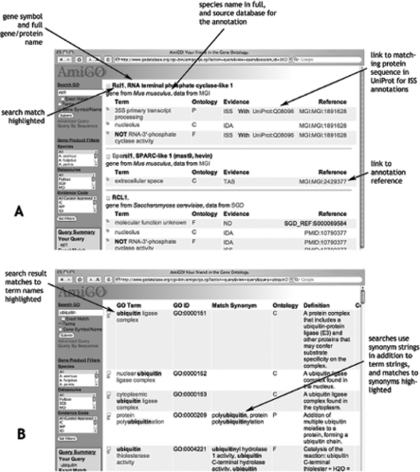
AmiGO (http://www.godatabase.org/cgi-bin/amigo/go.cgi) is a web resource developed by the GO Consortium for searching and browsing the Gene Ontology terms and gene product annotations. Recent enhancements include expanded searching of the ontology and gene products as well as improved display of search results. Synonyms, which may include phrases and terminology familiar to biologists and which clarify the meanings of GO terms, are now included in the GO term search and display. In addition, AmiGO now searches all available gene and gene product names provided by the annotation groups. The displays of search results and annotation data have been improved, as shown in Figure 1.
Figure 1.
Improved AmiGO interface. For all search results, the string matching the search query is highlighted to help identify why a specific result was returned. (A) The gene product search result display has been expanded to provide hyperlinks to documentation, references and other databases that contain information that support each annotation. Readability is greatly improved, such that for each search result, a sentence can be constructed from the tabular format, e.g. gene X from species S is annotated to term Y based on evidence of type Z from publications A, B and C. (B) The term search now includes comments; the search result display shows matches to synonyms as well as term names. Obsolete terms are grayed out and any suggested replacement terms are highlighted (data not shown).
APPLICATIONS OF GO DATA
In parallel with the growth of annotation coverage, GO's resources are now used in a number of different applications. The GO Bibliography, a collection of peer-reviewed literature on the development and usage of GO, has grown to over 600 publications (see http://www.geneontology.org/cgi-bin/biblio.cgi), documenting a number of novel uses of GO.
GO and gene expression
Among the most widespread applications of GO data is the use of GO terms and gene product annotations to help interpret the results of large-scale experiments, such as microarrays, in which any correlation between the functional information captured by GO and the expression patterns of a set of genes can help to highlight underlying biological phenomena. A large number of software tools have been developed to facilitate the analysis of gene expression data using GO (for a partial list see http://www.geneontology.org/GO.tools.microarray.shtml), and a paper reviewing the relative merits of a subset of these tools has recently been published (16).
GO terms and annotations have also been put to a variety of other uses, in both the biological and computer science communities. Biologists have used GO for tasks, such as gene function prediction (17), collaborative construction and analysis of cellular pathways (18), and association of genes to genetically inherited diseases (19). GO terms have also been incorporated into the Unified Medical Language System (UMLS) (20) maintained by the US National Library of Medicine (21). In the computer science community, GO has been used as a test of applying description logic (22,23) approaches to building sound, complete and logically consistent ontologies (22,24), and has featured in research into machine-processable ontologies (25) and into the automated checking of ontological consistency (26). Notably, GO terms offer a valuable standardized terminological resource to natural language processing researchers, facilitating information extraction from texts, knowledge discovery and ontology building from large collections of documents. For example, GO terms have been used in the Textpresso text mining system and in the BioCreAtIvE text mining assessment (13,27,28).
The NCI cancer biomedical informatics grid (caBIG) initiative and GO
The GO has been adopted by the caBIG initiative (https://cabig.nci.nih.gov/), enabling the cancer community to analyze microarray and proteomic data. Several available tools are now being integrated into caBIG, including GOminer (29,30), caArray, caWorkbench, RProteomics, Bioconductor (31), Reactome (32) and the cancer Pathways Interaction Database. In addition, caBIG has been integrating the Gene Ontology into the NCI Metathesaurus, Enterprise Vocabulary System and the cancer Data Standards Repository so that any caBIG project, dataset, or tool can take advantage of the GO. The GO has become the unifying terminology for the description and annotation of biological process, localization and function of gene products throughout the cancer research community.
ADDITIONAL ONTOLOGIES FOR BIOLOGY: OBO
The Gene Ontology is one of the ontologies held in the Open Biomedical Ontologies (OBO) collection (http://obo.sourceforge.net/). By providing controlled vocabularies that are freely available, OBO aims to extend GO development principles to many additional biological domains. There are currently over 40 ontologies lodged in OBO, covering domains such as anatomy, development, and phenotype, genomic and proteomic information and taxonomic classification. In addition to GO, OBO includes a relationship types ontology and the Sequence Ontology.
The OBO relationship types ontology (http://obo.sourceforge.net/relationship/) is an ontology of core relationship types, such as is_a, part_of, located_in and derives_from, with explicit definitions, to be used by all ontologies in the OBO collection (33).
SO: the Sequence Ontology
The Sequence Ontology (SO) provides terms and relationships for describing the features and attributes of biological sequences, e.g. DNA, RNA and proteins. Its purpose is to promote the standardization of sequence annotation among different organisms (34). The ontology currently contains 963 terms and 16 relationship types. A subset of the terms and relationships that describe located sequence features, known as SOFA (Sequence Ontology Feature Annotation), have been selected to provide a basic vocabulary to describe the products of automatic genome annotation efforts. SOFA is in its second release, and contains 179 terms. Ongoing development of both SO and SOFA proceeds via feedback and discussion from the annotation community through a mailing list and through soliciting the advice of domain experts.
The Sequence Ontology is primarily used to specify the type of annotation features in flat files [e.g. GFF3 (10)] and databases (e.g. CHADO, a relational database schema) (http://song.sourceforge.net/so_compliant_formats.shtml). SO and SOFA are currently being used to describe the annotations of several model organism genomes, both by automated pipeline and manual curation (http://song.sourceforge.net/so_groups.shtml). To facilitate integration with existing genome annotation projects, SO terms have been mapped to homologous terms in other biological vocabularies, such as the MGED ontology (35) and the GenBank feature table (36). These mappings are available on the web (http://song.sourceforge.net/so_mappings.shtml).
THE FUTURE OF GO
Ontology content
Work on immunology and on responses to stimuli is planned, and appropriate contacts are being made. The GO Consortium also hopes to tackle the areas of transport, signaling and neurobiology in the near future.
Gene product annotation
The GO Consortium will continue to update existing annotation datasets and work with new database groups that will annotate more species. In addition, curators and software developers will devise systems to enable bench scientists to contribute annotations for their domains of expertise.
Software/AmiGO
Further development and enhancement of AmiGO will make additional information about the organization of the ontology available and provide more up-to-date access to the annotations.
Acknowledgments
The Gene Ontology Consortium is supported by NIH/NHGRI grant HG02273 and has been supported by grants from the European Union RTD Programme ‘Quality of Life and Management of Living Resources’ (QLRI-CT-2001-00981 and QLRI-CT-2001-00015). Funding to pay the Open Access publication charges for this article was provided by NIH/NHGRI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gruber T.R. A translation approach to portable ontology specifications. Knowl. Acq. 1993;5:199–220. [Google Scholar]
- 2.Jones D.M., Paton R. Toward principles for the representation of hierarchical knowledge in formal ontologies. Data Knowl. Eng. 1999;31:99–113. [Google Scholar]
- 3.Stevens R., Goble C.A., Bechhofer S. Ontology-based knowledge representation for bioinformatics. Brief. Bioinform. 2000;1:398–414. doi: 10.1093/bib/1.4.398. [DOI] [PubMed] [Google Scholar]
- 4.Schulze-Kremer S. Ontologies for molecular biology and bioinformatics. In Silico Biol. 2002;2:179–193. [PubMed] [Google Scholar]
- 5.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Gene Ontology Consortium. Creating the Gene Ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blake J.A., Harris M.A. The Gene Ontology project: structured vocabularies for molecular biology and their application to genome and expression analysis. In: Baxevanis A.D., Davison D.B., Page R.D.M., Petsko G.A., Stein L.D., Stormo G., editors. Current Protocols in Bioinformatics. New York: John Wiley & Sons; 2003. [Google Scholar]
- 8.The Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris M.A., Lomax J., Ireland A., Clark J.I. The Gene Ontology project. In: Subramaniam S., editor. Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics. New York: John Wiley & Sons; 2005. [Google Scholar]
- 10.Lewis S.E. Gene Ontology: looking backwards and forwards. Genome Biol. 2005;6:103. doi: 10.1186/gb-2004-6-1-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eilbeck K., Lewis S.E. Sequence Ontology annotation guide. Comp. Funct. Genomics. 2004;5:642–647. doi: 10.1002/cfg.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolan M.E., Ni L., Camon E., Blake J.A. A procedure for assessing GO annotation consistency. Bioinformatics. 2005;21(Suppl. 1):i136–i143. doi: 10.1093/bioinformatics/bti1019. [DOI] [PubMed] [Google Scholar]
- 13.Camon E.B., Barrell D.G., Dimmer E.C., Lee V., Magrane M., Maslen J., Binns D., Apweiler R. An evaluation of GO annotation retrieval for BioCreAtIvE and GOA. BMC Bioinformatics. 2005;6(Suppl. 1):S17. doi: 10.1186/1471-2105-6-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camon E., Magrane M., Barrell D., Lee V., Dimmer E., Maslen J., Binns D., Harte N., Lopez R., Apweiler R. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mungall C.J. Obol: integrating language and meaning in bio-ontologies. Comp. Funct. Genomics. 2005;5:509–520. doi: 10.1002/cfg.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khatri P., Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King O.D., Foulger R.E., Dwight S.S., White J.V., Roth F.P. Predicting gene function from patterns of annotation. Genome Res. 2003;13:896–904. doi: 10.1101/gr.440803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demir E., Babur O., Dogrusoz U., Gursoy A., Ayaz A., Gulesir G., Nisanci G., Cetin-Atalay R. An ontology for collaborative construction and analysis of cellular pathways. Bioinformatics. 2004;20:349–356. doi: 10.1093/bioinformatics/btg416. [DOI] [PubMed] [Google Scholar]
- 19.Perez-Iratxeta C., Bork P., Andrade M.A. Association of genes to genetically inherited diseases using data mining. Nature Genet. 2002;31:316–319. doi: 10.1038/ng895. [DOI] [PubMed] [Google Scholar]
- 20.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lomax J., McCray A.T. Mapping the Gene Ontology into the unified medical language system. Comp. Funct. Genomics. 2004;4:345–361. doi: 10.1002/cfg.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevens R., Wroe C., Bechhofer S., Lord P., Rector A., Goble C. Building ontologies in DAML + OIL. Comp. Funct. Genomics. 2003;4:133–141. doi: 10.1002/cfg.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lord P., Stevens R.D., Goble C.A., Horrocks I. Description logics: OWL and DAML + OIL. In: Subramaniam S., editor. Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics. John Wiley & Sons; 2005. [Google Scholar]
- 24.Wroe C.J., Stevens R., Goble C.A., Ashburner M. A methodology to migrate the Gene Ontology to a description logic environment using DAML+OIL. Pac. Symp. Biocomput. 2003;2003:624–635. doi: 10.1142/9789812776303_0058. [DOI] [PubMed] [Google Scholar]
- 25.Williams J., Andersen W. Bringing ontology to the Gene Ontology. Comp. Funct. Genomics. 2003;4:90–93. doi: 10.1002/cfg.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh I., Karp P.D., Noy N.F., Altman R.B. Knowledge acquisition, consistency checking and concurrency control for Gene Ontology (GO) Bioinformatics. 2003;19:241–248. doi: 10.1093/bioinformatics/19.2.241. [DOI] [PubMed] [Google Scholar]
- 27.Hirschman L., Yeh A., Blaschke C., Valencia A. Overview of BioCreAtIvE: critical assessment of information extraction for biology. BMC Bioinformatics. 2005;6(Suppl. 1):S1. doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller H.-M., Kenny E.E., Sternberg P.W. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeeberg B.R., Feng W., Wang G., Wang M.D., Fojo A.T., Sunshine M., Narasimhan S., Kane D.W., Reinhold W.C., Lababidi S., et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeeberg B., Qin H., Narasimhan S., Sunshine M., Cao H., Kane D., Reimers M., Stephens R., Bryant D., Burt S., et al. High-Throughput GoMiner, an ‘industrial-strength’ integrative Gene Ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune deficiency (CVID) BMC Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith B., Ceusters W., Klagges B., Khler J., Kumar A., Lomax J., Mungall C., Neuhaus F., Rector A.L., Rosse C. Relations in biomedical ontologies. Genome Biol. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eilbeck K., Lewis S.E., Mungall C.J., Yandell M., Stein L., Durbin R., Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoeckert C.J., Parkinson H. The MGED ontology: a framework for describing functional genomics experiments. Comp. Funct. Genomics. 2003;4:127–132. doi: 10.1002/cfg.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]