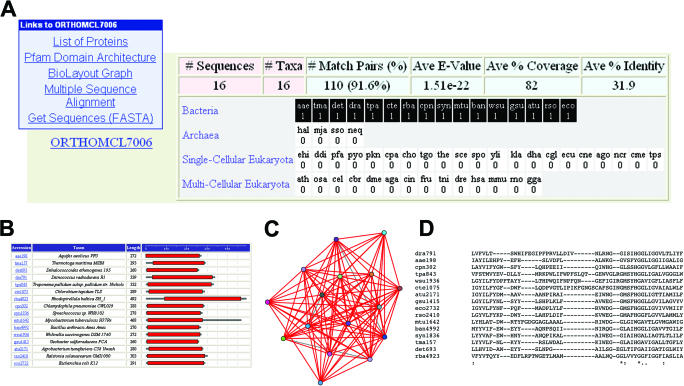
Figure 2.
An OrthoMCL group is a cluster of sequences from multiple species predicted to be orthologous to each other. (A) Ortholog group summary information, including group size (# Sequences, # Taxa), BLAST statistics (% Match Pairs, Average E-value, Average % Coverage, Average % Identity) and the phyletic pattern profile for all species in the dataset is shown. Rows in the phyletic pattern profile table represent bacteria, archaea, single-cellular eukaryotes and multi-cellular eukaryotes (plants and animals); each box represents a single species, with black or white background denoting presence or absence in the ortholog group, and the number of protein sequences found in the ortholog group listed. Mouse-over expands abbreviations to provide the full species name. Links at top left access a tabular list of information for each member of the ortholog group (including links to the reference database), a graphical representation of Pfam domain architecture (B), a BioLayout graph of pairwise similarity scores (C), a MUSCLE multiple sequence alignment (D) and a sequence retrieval option. The example shown illustrates a ‘prolipoprotein diacylglyceryl transferase’, whose distribution is restricted to the bacteria.