Abstract
The arctic flora is considered to be impoverished, but estimates of species diversity are based on morphological assessments, which may not provide accurate counts of biological species. Here we report on crossing relationships within three diploid circumpolar plant species in the genus Draba (Brassicaceae). Although 99% of parental individuals were fully fertile, the fertility of intraspecific crosses was surprisingly low. Hybrids from crosses within populations were mostly fertile (63%), but only 8% of the hybrids from crosses within and among geographic regions (Alaska, Greenland, Svalbard, and Norway) were fertile. The frequent occurrence of intraspecific crossing barriers is not accompanied by significant morphological or ecological differentiation, indicating that numerous cryptic biological species have arisen within each taxonomic species despite their recent (Pleistocene) origin.
Keywords: crossing relationships, cryptic species, Draba, speciation, postmating isolation
The Arctic flora has long been viewed as depauperate. Indeed, the decrease in biological diversity with increasing latitude is one of the oldest recognized patterns in ecology (1). Diversity has, however, typically been quantified as the number of morphological or “taxonomic” species. Little is known about biological species diversity in the Arctic (2), because the recognition of biological species requires information on reproductive isolation.
Here we present results from crossing experiments within three diploid circumpolar plant species: Draba fladnizensis, Draba nivalis, and Draba subcapitata (Table 1, which is published as supporting information on the PNAS web site). Low levels of genetic differentiation within and among the species suggest they originated recently, probably within the last one million years (3). The species are mainly self-pollinated (4), but they occasionally outcross as demonstrated by reports of natural hybrids between D. fladnizensis and D. nivalis (5). We show that, contrary to expectations based on observations of limited morphological, ecological, and genetic diversity, numerous cryptic biological species have arisen within each of the taxonomic species studied here. These results imply that biological species diversity may be considerably higher in arctic regions than previously believed.
Results
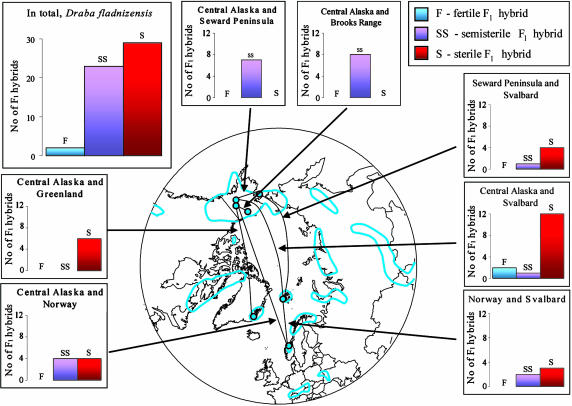
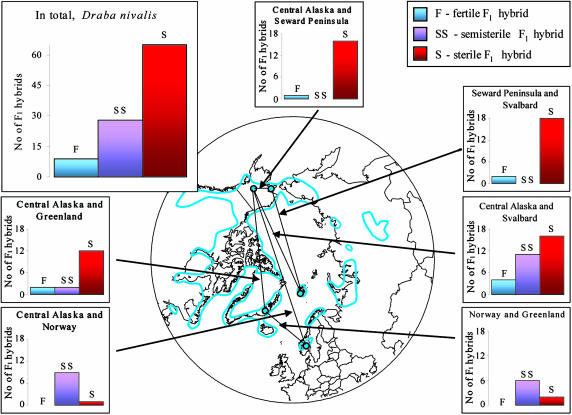
All of the 79 parental plants of D. fladnizensis and D. nivalis and six of the seven parental plants of D. subcapitata were highly fertile (Table 1). The within-population crosses mostly generated fertile hybrids (10 of 16 crosses attempted), but six crosses resulted in semisterile hybrids (Tables 2 and 3, which are published as supporting information on the PNAS web site). The within-region crosses (among populations in Alaska) generated hybrids that were semisterile in D. fladnizensis (Fig. 1 and Tables 2 and 3) and mostly sterile in D. nivalis (Fig. 2 and Tables 2 and 3). Likewise, the majority of crosses among regions resulted in sterile F1 hybrids (73% in D. fladnizensis, 58% in D. nivalis, and 50% in D. subcapitata; Tables 2 and 3). The among-region crosses resulted in only a handful of fertile hybrids (5% in D. fladnizensis and 9% in D. nivalis).
Fig. 1.
Crossing relationships within D. fladnizensis, summarized as results of within-region crosses (Alaska: Central Alaska, Seward Peninsula, and Brooks Range) and among-region crosses (Alaska, Greenland, Svalbard, and Norway; combinations represented by less than five crosses are omitted). The blue line delimits the total geographic distribution of the species; blue dots show the origins of material used in this study.
Fig. 2.
Crossing relationships within D. nivalis, summarized as results of within-region crosses (Alaska: Central Alaska and Seward Peninsula) and among-region crosses (Alaska, Greenland, Svalbard, and Norway; combinations represented by less than five crosses are omitted). The blue line delimits the total geographic distribution of the species; blue dots show the origins of material used in this study.
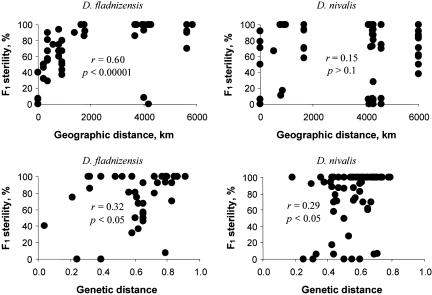
Correlations between geographic and genetic amplified fragment length polymorphism (AFLP) distances were significant and fairly strong (Mantel's test, r = 0.49 for D. fladnizensis and r = 0.41 for D. nivalis; P < 0.00001). Likewise, weaker but significant correlations were observed between F1 sterility and genetic distance in both D. fladnizensis (r = 0.32) and D. nivalis (r = 0.29) (Fig. 3). Geographic distance was strongly correlated with F1 sterility (r = 0.60) in D. fladnizensis but not in D. nivalis (Fig. 3).
Fig. 3.
Hybrid sterility and geographic/genetic distances. Association between F1 hybrid sterility (as inverse seed set) and the geographic and genetic AFLP distances between their parental populations. Genetic distances are given as inverse Jaccard's coefficients.
Discussion
Our results indicate that numerous cryptic biological species exist within D. fladnizensis and D. nivalis; 92% of the within- and among-region crosses resulted in sterile or semisterile hybrids despite the full fertility of parental plants. Even crosses within populations yielded some semisterile hybrids, implying that hybrid incompatibilities arise extremely rapidly in these arctic Draba species. The few data for D. subcapitata suggest a similar pattern for this diploid species as well.
So why are postzygotic isolation barriers accumulating so quickly within these species and on such a small geographic scale? In other plant species, the development of such barriers has been shown to be associated with morphological (6–8) and/or ecological divergence (9), as might be predicted if hybrid incompatibility was a byproduct of divergent ecological selection. However, ecological selection seems unlikely to account for the rapid evolution of crossing barriers in these Draba species because they appear morphologically and ecologically homogeneous. Likewise, sexual selection, which partially explains the rapid evolution of hybrid male sterility in animals (10), cannot account for the accelerated accumulation of incompatibilities in hermaphroditic plants.
The most plausible explanation for our findings relates to the selfing mating system of all three species. Selfing, which is considered to be frequent in arctic plants (11), provides instantaneous isolation from other lineages, thereby facilitating the accumulation of hybrid incompatibilities. Inbreeding also greatly increases the fixation rate of underdominant mutations through drift, whether they have a chromosomal or genic basis. Finally, postzygotic barriers could arise as a byproduct of compensatory mutations that ameliorate the negative effects of inbreeding (12).
Because regular meiosis has been observed in sterile, intraspecific D. fladnizensis hybrids (13), major chromosomal differences seem unlikely to account for the sterility barriers in these diploid Draba, although microchromosomal rearrangements cannot be ruled out. The correlation observed in this study between genetic distance and hybrid sterility further implies that postzygotic barriers accumulate in a gradual (clock-like) fashion in this group and may therefore involve many loci (14, 15).
Cryptic biological species are commonly detected in temperate lineages of selfing plant species as well (16) but not on the scale observed here. Also, many of the well studied temperate selfers exhibit high rates of chromosomal evolution (9, 17) that are not observed in these species of Draba. Possibly, a greater diversity of cryptic biological species in selfing arctic lineages relative to their temperate counterparts relates to lineage age. Selfing temperate species may be of short duration because of competition with outcrossing congeners or newly arisen selfers that lack accumulated load (18). Outcrossing species appear to be less frequent in the Arctic (11), however, which may reduce the rate of origin and turnover of selfing lineages.
Although the recent speciation literature has appropriately emphasized the role of selection in the development of reproductive isolation (19–22), the present study provides a possible counterexample; because of selfing, genetic drift could provide an efficient means for the accumulation of hybrid incompatibilities.
In conclusion, although the Arctic is comparatively poor in morphological species, it may be rich in cryptic, biological species as demonstrated here for three species of Draba. An important unanswered question is whether the results from Draba can be extrapolated to other arctic plants or animals.
Methods
Experimental Crosses. Fifteen populations (34 plants) of D. fladnizensis, sixteen populations (45 plants) of D. nivalis, and four populations (7 plants) of D. subcapitata were used in the crossing experiments (Table 1). Ploidy and taxonomic assignment of each population were verified by chromosome number determinations, flow cytometry, and molecular analyses (3, 23, 24). The plants were cultivated in an insect-free phytotrone at the University of Oslo (cultivation conditions as specified in ref. 25). Because the spatial structure of potential isolating barriers initially was unknown, we made crosses within populations, within regions (Alaska), and among regions (Alaska, Greenland, Svalbard, and Norway). A comparatively low number of crosses were made within populations because incompatibility was expected to be more frequent among geographically more distant plants. In all (replicates yielding identical results were omitted; see Table 2), 6 within-population, 23 within-region, and 41 among-region crosses were made for D. fladnizensis; 9 within-population, 19 within-region, and 85 among-region crosses were made for D. nivalis; and 1 within-population and 4 among-region crosses were made for D. subcapitata (Tables 2 and 3). Flower buds of D. subcapitata are very small and difficult to emasculate, which accounts for the more limited results obtained for this species.
In each crossing experiment, the anthers were removed from 1–4 flower buds of one maternal plant before anthesis, and pollen was transferred 1–3 days later. The seeds were stored for at least 3 months at 4°C, scarified with fine sandpaper, and sowed. Three F1 seedlings (if available) from each cross were raised to maturity. One randomly chosen F1 hybrid from each cross was analyzed in detail, but all three F1 hybrids were briefly examined for seed set. F1 hybrid fertility was estimated as percent viable pollen (% pollen stainability in cotton blue) and as percent seed set after spontaneous selfing (% developed seeds of total number of ovules). It has previously been shown that, in F1 hybrids of Draba, seed set does not increase after hand-selfing with excess self-pollen compared with that after spontaneous selfing (13). Because pollen stainability may vary during the course of flowering, additional samples were taken if the stainability was low, i.e., <80%. If more than one pollen or seed sample was taken for a plant, the one showing the highest fertility was used. All parental plants used in the crossing experiments were also tested for fertility as described above and served as controls for the within- and among-population crosses. Seed set and pollen viability were highly correlated (Pearson's r = 0.9, P < 0.0001, n = 188), which made it possible to classify the hybrids as fertile (average pollen viability and seed set ≥80%, i.e., equivalent to the parental plants), semisterile (pollen viability and seed set <80% and seed set >0%), or sterile (pollen viability <10% and seed set = 0%).
Molecular Marker Surveys. To determine whether reductions in hybrid fertility were associated with genetic and/or geographic divergence among populations, we analyzed 43 and 46 plants of D. fladnizensis and D. nivalis, respectively, for AFLPs (26). The plants chosen for AFLP analyses were derived from the same populations (often the same plants) used in the crossing experiments (Table 1). Approximately 75% of the plants used in the crossings were also analyzed for AFLPs, and additional plants from the same populations were also included.
DNA was isolated after the cetyltrimethylammonium bromide (CTAB) method (27) with an additional purifying RNase step (28) or by using the DNeasy kit according to the DNeasy plant mini handbook (Qiagen, Valencia, CA). DNA was extracted from 15–30 mg of silica-dried leaves and 20–50 mg (CTAB) or 100 mg (DNeasy kit) of fresh leaves. Total genomic DNA was digested with the restriction enzymes MseI and EcoRI according to the AFLP plant mapping kit protocol (Applied Biosystems). Two geographically distant plants of each species were used in a primer test, and 3 of the 40 primer pairs tested were selected for full analysis (E-ACT/M-CTA, E-AGG/E-CTT, and E-ACC/E-CTT). The AFLP markers were visualized with genescan analysis 3.1 software (Applied Biosystems). Fluorescence peaks were inspected manually, and distinct, polymorphic peaks in the 60- to 500-bp range were scored as present or absent. The two species were scored independently of each other, producing two AFLP data sets.
Pairwise genetic AFLP distances were calculated as the inverse of Jaccard's similarities. Pairwise geographic distances between populations were calculated by using the program inverse from the National Geodetic Survey (www.ngs.noaa.gov/PC_PROD/Inv_Fwd). inverse computes the geodetic azimuth and ellipsoidal distance between two points based on their locations in latitudes and longitudes, thus accounting for the curvature of the earth. The reference ellipsoid WGS84 was chosen. Correlations between geographic and genetic AFLP distances were calculated for each species by using Mantel's test in NTSYSpc (29). Pearson's correlation coefficients were calculated in past (30) for pairwise comparisons between geographic distance and hybrid sterility and between genetic distance and hybrid sterility for each species.
Supplementary Material
Acknowledgments
We thank the collectors of material (see notes in Table 1), especially Reidar Elven for collections in Alaska and Anne-Cathrine Scheen for providing material from Svalbard. We also thank Erlend Råheim for advice regarding computation of geographic distances, Inger Greve Alsos for help with preparation of figures, and the gardeners at the Department of Biology and in the Botanical Garden at the University of Oslo for taking care of the cultivated plants. This work was supported by Research Council of Norway Grants 123714/410 and 146515/420 (to C.B.).
Conflict of interest statement: No conflicts declared.
Abbreviation: AFLP, amplified fragment length polymorphism.
References
- 1.Willig, M. R., Kaufman, D. M. & Stevens, R. D. (2003) Annu. Rev. Ecol. Evol. Syst. 34, 273–309. [Google Scholar]
- 2.Gaston, K. J. (2000) Nature 405, 220–227. [DOI] [PubMed] [Google Scholar]
- 3.Grundt, H. H., Popp, M., Brochmann, C. & Oxelman, B. (2004) Mol. Phylogenet. Evol. 32, 695–710. [DOI] [PubMed] [Google Scholar]
- 4.Brochmann, C. (1993) Plant Syst. Evol. 185, 55–83. [Google Scholar]
- 5.Ekman, E. (1932) Sven. Bot. Tidskr. 26, 198–200. [Google Scholar]
- 6.Grun, P. (1961) Am. J. Bot. 48, 79–89. [Google Scholar]
- 7.Tai, W. & Vickery, R. K., Jr. (1970) Evolution (Lawrence, Kans.) 24, 670–679. [DOI] [PubMed] [Google Scholar]
- 8.Mooring, J. S. (2001) Am. J. Bot. 88, 285–312. [PubMed] [Google Scholar]
- 9.Snyder, L. A. (1950) Am. J. Bot. 37, 628–636. [Google Scholar]
- 10.Wu, C.-I. (1992) Evolution (Lawrence, Kans.) 46, 1584–1587. [Google Scholar]
- 11.Lloyd, D. G. (1980) in Demography and Evolution in Plant Populations, ed. Solbrig, O. T. (Blackwell Scientific, Oxford), pp. 67–88.
- 12.Estes, S. & Lynch, M. (2003) Evolution (Lawrence, Kans.) 57, 1022–1030. [DOI] [PubMed] [Google Scholar]
- 13.Brochmann, C., Borgen, L. & Stedje, B. (1992) Nord. J. Bot. 13, 121–147. [Google Scholar]
- 14.Presgraves, D. C. (2002) Evolution (Lawrence, Kans.) 56, 1168–1183. [DOI] [PubMed] [Google Scholar]
- 15.Moyle, L. C., Olson, M. S. & Tiffin, P. (2004) Evolution (Lawrence, Kans.) 58, 1195–1208. [DOI] [PubMed] [Google Scholar]
- 16.Grant, V. (1981) Plant Speciation (Columbia Univ. Press, New York), 2nd Ed.
- 17.Grant, V. (1964) Adv. Genet. 12, 281–328. [Google Scholar]
- 18.Lynch, M., Conery, J. & Burger, R. (1995) Evolution (Lawrence, Kans.) 49, 1067–1080. [DOI] [PubMed] [Google Scholar]
- 19.Macnair, M. R., Macnair, V. E. & Martin, B. E. (1989) New Phytol. 112, 269–279. [Google Scholar]
- 20.Barton, N. H. (1996) Philos. Trans. R. Soc. London B 351, 785–795. [DOI] [PubMed] [Google Scholar]
- 21.Noor, M. A. F. (2003) Nature 423, 699–700. [DOI] [PubMed] [Google Scholar]
- 22.Presgraves, D. C., Balagopalan, L., Abmayr, S. M. & Orr, H. A. (2003) Nature 423, 715–719. [DOI] [PubMed] [Google Scholar]
- 23.Scheen, A.-C., Elven, R. & Brochmann, C. (2002) Can. J. Bot. 80, 59–71. [Google Scholar]
- 24.Grundt, H. H., Obermayer, R. & Borgen, L. (2005) Bot. J. Linn. Soc. 147, 333–347. [Google Scholar]
- 25.Brochmann, C., Soltis, P. S. & Soltis, D. E. (1992) Nord. J. Bot. 12, 257–272. [Google Scholar]
- 26.Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M. & Zabeau, M. (1995) Nucleic Acids Res. 23, 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hombergen, E. J. & Bachmann, K. (1995) Theor. Appl. Genet. 90, 853–858. [DOI] [PubMed] [Google Scholar]
- 28.Gabrielsen, T. M. & Brochmann, C. (1998) Mol. Ecol. 7, 1701–1708. [Google Scholar]
- 29.Rohlf, F. (1999) Numerical Taxonomy and Multivariate Analysis System (Exeter Software, New York), Version 2.02.
- 30.Hammer, Ø., Harper, D. A. T. & Ryan, P. D. (2001) Palaeontologia Electronica 4 (1), 9 pp. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.