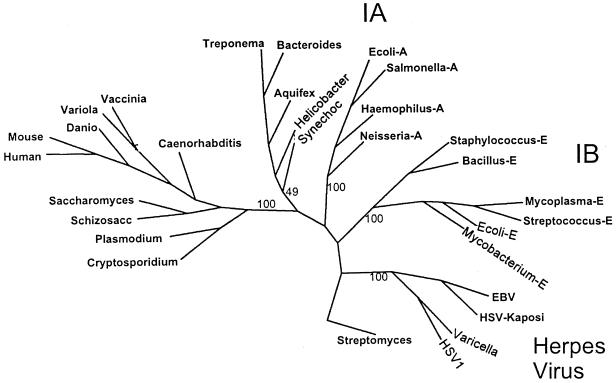
FIG. 3.
Unrooted phylogenetic tree showing the relationship of B. fragilis NrdA to representative orthologs of class I RRase large subunits. A 148-amino-acid internal region of the peptides was aligned using the PILEUP program from the Wisconsin GCG software package. The parsimony program (Protpars) was used to generate the consensus tree from 100 bootstrap analyses, but not all species used in the alignment are shown on the tree for the sake of clarity. The bootstrap values for the major branches of the tree are shown and the Streptomyces clavuligerus class II enzyme (labeled Streptomyces) was used as the outgroup. Abbreviations: Mycoplasma-E, Mycoplasma genitalium; Streptococcus-E, Streptococcus dysgalactiae; Mycobacterium-E, Mycobacterium tuberculosis; Ecoli-E, E. coli nrdE, Salmonella-A, S. enterica serovar Typhimurium nrdA; Bacillus-E, Bacillus subtilis; Haemophilus-A, Haemophilus influenzae; Neisseria-A, Neisseria meningitidis; Ecoli-A, E. coli nrdA, mouse, Mus musculus; human, Homo sapiens; Danio, Danio rerio; vaccinia, vaccinia virus; variola, variola virus; Caenorhabditis, Caenorhabditis elegans; Schizosacc, Schizosaccharomyces pombe; Saccharomyces, Saccharomyces cerevisiae; Plasmodium, Plasmodium falciparum; Cryptosporidium, Cryptosporidium parvum; Synechoc, Synechocystis spp.; Bacteroides, Bacteroides fragilis; Aquifex, Aquifex aeolicus; Helicobacter, Helicobacter pylori; Treponema, Treponema pallidum, HSV-Kaposi, Kaposi's sarcoma-associated herpesvirus; EBV, Epstein-Barr virus; varicella, varicella-zoster virus; HSV1, human herpes simplex virus type 1.