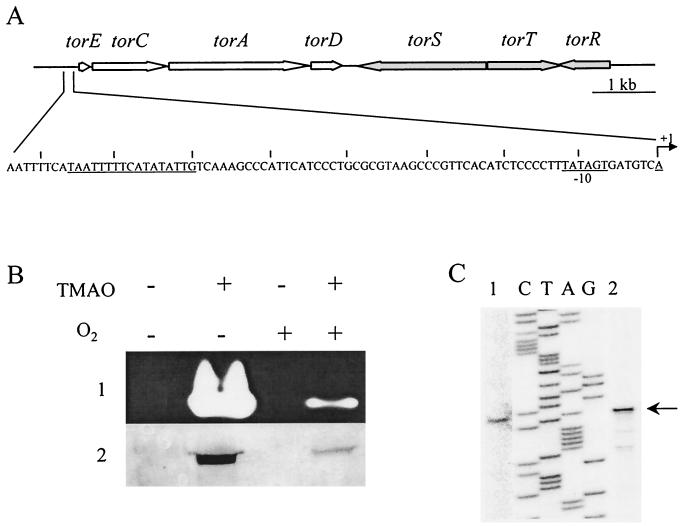
FIG. 1.
(A) Physical map of the tor locus of S. oneidensis. The large arrows show the locations and orientations of the tor genes. The nucleotide sequence of the torECAD regulatory region is indicated. The +1 arrow corresponds to the transcription start site. The putative −10 promoter box and the TorR binding region are underlined. (B) SDS-PAGE analysis of TMAO reductase production of S. oneidensis grown anaerobically (−O2) or aerobically (+O2), in the presence (+) or absence (−) of TMAO. Nonheated (row 1) or heated (row 2) periplasmic extracts (15 μg) were loaded on the gel. After electrophoresis, the presence of TorAso was checked by staining the gel for TMAO reductase activity (row 1) or by a Western blot with anti-TorAsm antibodies (row 2). (C) Localization of the transcription start site of torE. A labeled primer complementary to torE internal sequence was annealed to total RNA from S. oneidensis cells grown anaerobically in the presence of TMAO (lane 1) or to RNA from E. coli LCB436 (pPTor4-pRso2) cells grown anaerobically in the presence of 0.001% arabinose (lane 2), and extended with reverse transcriptase. The primer extension reactions were loaded on each side of a sequencing ladder of the torE region made with the same primer as in the extension reaction.