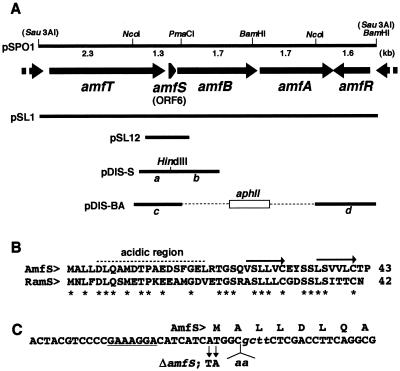
FIG. 1.
Restriction map of the 9-kb Sau3AI fragment and the positions and directions of ORFs in the fragment (A), amino acid alignment of AmfS and the homolog in S. coelicolor (B), and nucleotide sequence of a part of amfS (C). (A) The extents and directions of ORFs of the nucleotide sequence predicted by the FRAME program (2) are indicated by arrows. The positions of fragments used for gene disruption and complementation experiments are also shown. pSL1 and pSPO1 (23) carry the 9-kb fragment on a low-copy-number plasmid, pIJ922 (8), and a high-copy-number plasmid, pIJ487 (27), respectively. pSL12 carries the amfS-containing 700-bp region at the BamHI site of pIJ922. (B) The amino acid sequence of AmfS was aligned with the homologous sequence identified in S. coelicolor A3(2). The C-terminal hydrophobic repeats are indicated by arrows. Idential amino acids are indicated by asterisks. (C) Nucleotide sequence of the N-terminal portion of amfS with the deduced amino acid sequence. The potential site for ribosome binding is underlined. The mutated sequence used for the disruption of amfS (ΔamfS) is indicated below the wild-type sequence. The italic lowercase letters represent the recognition site for HindIII created to check for true recombination.