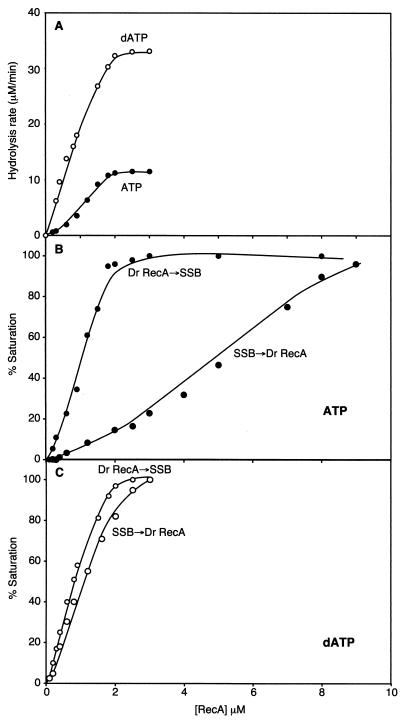
FIG. 4.
Effect of native RecADr protein concentration on the rate of (d)ATP hydrolysis. (A) Reactions were carried out as described in Materials and Methods, and reaction mixtures contained 6 μM circular single-stranded φX174 DNA, 0.6 μM E. coli SSB, 2 mM ATP (or dATP), and the indicated concentration of native RecADr protein. Reactions carried out with the two different nucleotide cofactors are identified in the figure. (B) Order of addition effects in the presence of ATP. Reactions were carried out as for panel A, with 2 mM ATP. RecA protein was added to ssDNA, which was followed by addition of ATP and SSB (RecADr→SSB reactions), or SSB protein was added to ssDNA, which was followed by addition of ATP and RecA protein (SSB→RecADr reactions). (C) Reactions were carried out as for panel B, but with dATP replacing ATP.