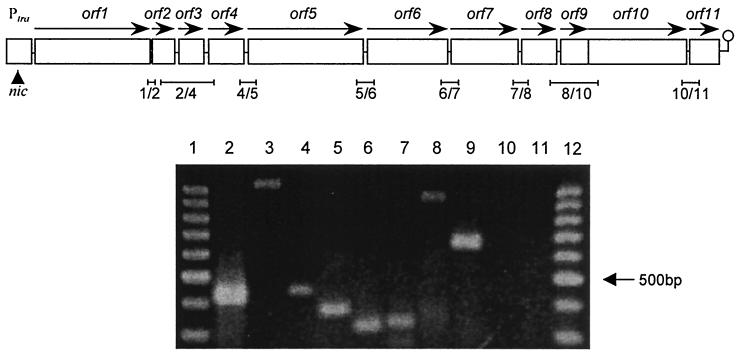
FIG. 2.
(Top) Organization of the tra region encompassing orf1 to orf11. Open reading frames are shown as boxes with horizontal arrows indicating the direction of transcription. The oriT nick site is indicated by an arrowhead. RT-PCR products are shown below the diagram. They are named according to the genes amplified, counting from orf1 to orf11. (Bottom) Cotranscription of the tra genes. Except for the amplification of the products from orf2 to orf4 and orf8 to orf10, the conditions were as follows. Incubation at 48°C for 40 min for RT was followed by inactivation of avian myeloblastosis virus reverse transcriptase and denaturation of the template at 94°C for 90 s. A cycle program consisting of 94°C for 30 s, 60°C for 90 s, and 68°C for 2 min was then applied for 30 cycles and terminated by a final elongation step of 7 min. For the amplification of products from orf2 to orf4 and orf8 to orf10, the RT step lasted 60 min and a cycle program consisting of 94°C for 30 s, 55°C for 90 s (51°C for orf8 to orf10), and 68°C for 4 min was applied for 35 cycles and terminated by a final elongation step of 8 min. The RT-PCR products were loaded onto 2.2% Tris-acetate-EDTA gels. Lanes 1 and 12, 100-bp DNA ladder (Promega); lane 2, product from orf1 and -2 (expected size, 406 bp); lane 3, product from orf2 to -4 (expected size, 1,035 bp); lane 4, product from orf4 and -5 (expected size, 433 bp); lane 5, product from orf5 and -6 (expected size, 359 bp); lane 6, product from orf6 and -7 (expected size, 322 bp); lane 7, product from orf7 and -8 (expected size, 326 bp); lane 8, product from orf8 to -10 (expected size, 932 bp); lane 9, product from orf10 and -11 (expected size, 690 bp). No detectable products were obtained in control reactions with each pair of primers with RNA from the plasmid-free isogenic strain E. faecalis JH2-2 (lane 10, with primers for orf1 and orf2) and with RNA from E. faecalis JH2-2(pIP501) with the RT step omitted (lane 11, with primers for orf1 and orf2).