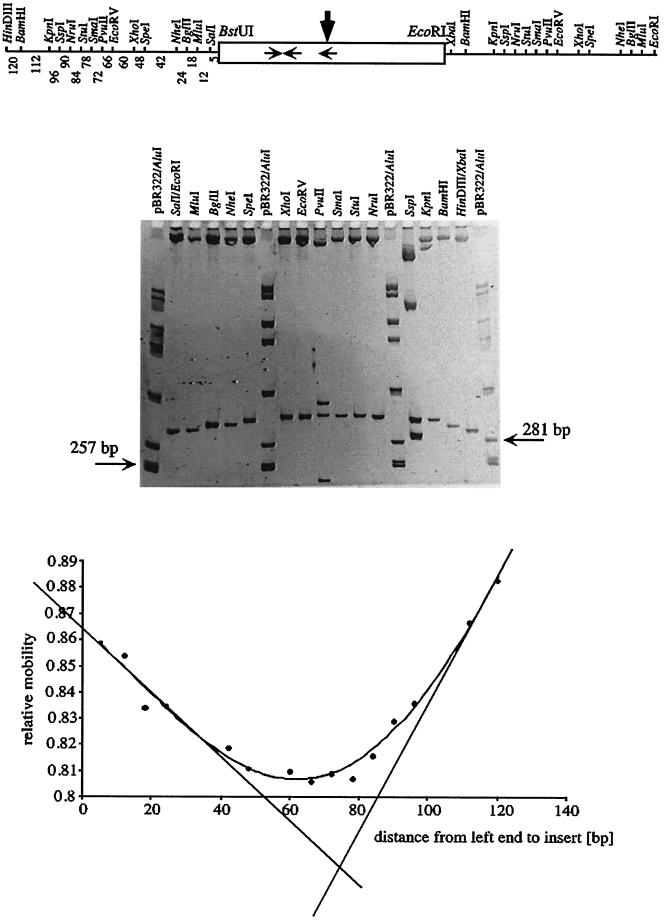
FIG. 7.
Determination of DNA bending within the 128-bp BstUI-EcoRI fragment from the pcaU-pcaI intergenic region (Fig. 1) by a circular permutation assay using bending vector pBend5. (Top) The relevant part of plasmid p5/128 is shown with the restriction sites of the permutation element and the test DNA as a box in the middle. Horizontal arrows, locations of the PcaU binding sites; vertical arrow, bending center revealed in this experiment. Numbers under the left half of the permutation element, distances in base pairs between the BstUI site of the test DNA and the left end of the fragment resulting from restriction digestion with the respective enzymes. These numbers were used as the distances from left end to insert in the graph (bottom). (Middle) Example of a gel containing samples of plasmid p5/128 after restriction cleavage with the enzymes indicated. The permutated fragments had a length of 255 bp; for comparison two fragments of the standard are indicated. (Bottom) Plot of the relative mobilities of the fragments against the distance between the left end of the permutated fragments and the test DNA enables the determination of the bending center. Relative mobilities are the averages of six gels; the error was between 0.7 and 1.5%.