Abstract
The sol operon of Clostridium acetobutylicum is the essential transcription unit for formation of the solvents butanol and acetone. The recent proposal that transcriptional regulation of this operon is controlled by the repressor Orf5/SolR (R. V. Nair, E. M. Green, D. E. Watson, G. N. Bennett, and E. T. Papoutsakis, J. Bacteriol. 181:319-330, 1999) was found to be incorrect. Instead, regulation depends on activation, most probably by the multivalent transcription factor Spo0A. The operon is transcribed from a single promoter. A second signal identified in primer extension studies results from mRNA processing and can be observed only in the natural host, not in a heterologous host. The first structural gene in the operon (adhE, encoding a bifunctional butyraldehyde/butanol dehydrogenase) is translated into two different proteins, the mature AdhE enzyme and the separate butanol dehydrogenase domain. The promoter of the sol operon is preceded by three imperfect repeats and a putative Spo0A-binding motif, which partially overlaps with repeat 3 (R3). Reporter gene analysis performed with the lacZ gene of Thermoanaerobacterium thermosulfurigenes and targeted mutations of the regulatory region revealed that the putative Spo0A-binding motif, R3, and R1 are essential for control. The data obtained also indicate that an additional activator protein is involved.
Regulation of butanol formation by the obligately anaerobic bacterium Clostridium acetobutylicum, the model organism used in molecular biology for the apathogenic clostridia, is still not completely understood. Research during the last decade led to cloning and sequencing of the sol operon, designated on the basis of its function in solvent formation, which includes the genes that encode a butyraldehyde/butanol dehydrogenase (adhE) and an acetoacetyl coenzyme A:butyrate/acetate coenzyme A transferase (ctfA and ctfB) (9, 21). This operon is located on a megaplasmid (6, 7), and transcriptional induction of it marks the onset of butanol formation (30). During the solvent production phase, the sol operon is shut down, and the monocistronic bdhB operon, which encodes another butanol dehydrogenase and is located on the chromosome, takes over after it is induced (27, 30). The corresponding butyraldehyde dehydrogenase is still unknown. It has been proposed that the sol operon is controlled by two promoters, based on primer extension studies (9, 21). The sequence of the putative distal promoter, designated P1 or S2, exhibited almost perfect homology to the consensus sequence of housekeeping promoters (just one mismatch), whereas the sequence of the putative proximal promoter (P2 or S1) had at least five mismatches (in the 12 nucleotides comprising the −35 and −10 regions) and unusual spacing of the two promoter boxes (8, 9, 21). Nevertheless, based on signal intensities obtained in primer extension experiments, most of the transcripts are initiated at this transcription start point (9).
A major step forward in understanding the regulation described above seemed to be the recent report that an open reading frame (orf5) located directly upstream of the sol operon encodes a transcriptional repressor for this locus (22). This report was based on the following findings: (i) overexpression of Orf5 (in that report designated SolR) resulted in a solvent-negative phenotype, (ii) insertional inactivation of the orf5 gene led to mutants with markedly improved solvent yields, and (iii) a potential helix-turn-helix DNA-binding motif was identified within the Orf5 protein (22). However, contradictory data were also reported. Purified Orf5 did not bind to the sol operon regulatory region when it was tested in gel retardation assays with either linear or supercoiled DNA templates (33). In addition, this protein was found to be localized on the extracellular side of the cytoplasmic membrane, to be involved in glycosylation-deglycosylation reactions, and to contain a tetratrico peptide repeat protein-protein interaction motif instead of a helix-turn-helix sequence (33). Most strikingly, the solvent-negative phenotype observed upon overexpression could not be reproduced. These mutants even produced 15% more butanol than the wild type (33). These findings clearly rule out the possibility that Orf5 is a transcriptional repressor of the sol operon. While insertional inactivation of the orf5 gene could have a (possibly secondary) effect on solventogenesis by affecting the glycosylation-deglycosylation activity in the cell, the contradictory reports concerning solvent production and nonproduction in Orf5-overexpressing strains have remained mysterious.
In this paper, we describe a detailed analysis of the sol operon regulatory region and provide evidence that the reported solvent-negative phenotype (22) results from erroneous subcloning of part of the regulatory region of the sol operon together with the orf5 gene. This DNA fragment carries a putative binding motif for the multivalent transcription factor Spo0A, which is required for transcriptional induction.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this investigation are listed in Table 1. Escherichia coli WL3(pGP1-2) was used for heterologous overexpression of His6-tagged AdhE from pTWa4-2::adhE6×His, and in vivo plasmid methylation was carried out in E. coli ER2275(pAN1) (20). For all other procedures E. coli XL1-B was used. All strains were routinely grown in Luria-Bertani medium for both DNA and protein preparation. C. acetobutylicum DSM 792 was grown anaerobically under an N2 atmosphere in 2× YT medium (26); for reporter protein expression analyses cultivation was carried out in morpholineethanesulfonic acid (MES)-buffered minimal medium (3). When necessary, media were supplemented with ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), clarithromycin (5 μg/ml), or erythromycin (50 μg/ml).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description or genotype | Source or reference |
|---|---|---|
| Escherichia coli | ||
| ER2275 | trp-31 his-1 tonA2 rpsL104 supE44 xyl-7 mtl-2 metB1 e14− Δ(lac)U169 endA1 recA1 R(zbgZ10::Tn10) Tcs Δ(mcr-hsd-mrr)114::1510 F′ [proAB traD36 laq1qΔM15 zzf::mini Tn10 (Kmr)] | 20 |
| WL3 | adhC81 fadR adhE3 | 17 |
| XL1-B | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 [F′ proAB lac1qZΔM15 Tn10 (Tcr)] | Stratagene GmbH, Heidelberg, Germany |
| Clostridium acetobutylicum DSM 792 | Wild type | DSMZ, Braunschweig, Germanya |
| Plasmids | ||
| pACT1 | C terminus orf5 and intergenic region between orf5 and P1sol in pIMP1 | This study |
| pAN1 | Cmr, φ3tI, p15A oriR | 20 |
| pGP1-2 | Tn903 (Kmr), cI857, T7 polymerase under control of λPL, P15A oriR | 32 |
| pIMP1 | Emr (ermC) Apr, pMB1 oriR, pIM13 oriR(+) | 19 |
| pK9 | Apr, ColE1 oriR, rop, harboring sol and orf5 | 9 |
| pKLIMP7 | P1-His6-tagged adhE fusion in pIMP1 | 23 |
| pKLIMP12 | P2-[ΔP1]-His6-tag-encoding adhE fusion in pIMP1 | 23 |
| pKLIMP17 | P1-[Δ hairpins upstream of the adhE start codon]-His6-tag-encoding adhE fusion in pIMP1 | 23 |
| pLacZF | lacZ from T. thermosulfurigenes in pIMP1 | This study |
| pSN51 | Apr, tetM, pMB1 oriR, pAMβ1 oriR(+), oriT, Padc-lacZ-fusion | 23 |
| pTWa4-2 | Apr, ColE1 oriR, f1 ori, T7 promoter, atpE TIR | 36 |
| pTWa4-2::adhE6×His | His6-tag-encoding adhE in pTWa4-2 | This study |
| pZF-sol | P1-lacZ fusion in pLacZF | This study |
| pZF-sol-act | P1-[Δ activator region]-lacZ fusion in pLacZF | This study |
| pZF-sol-0A | P1-[mutated Spo0A-binding site]-lacZ fusion in pLacZF | This study |
| pZF-sol-R1 | P1-[mutated activator region]-lacZ fusion in pLacZF | This study |
| pZF-sol-R2 | P1 [mutated activator region]-lacZ fusion in pLacZF | This study |
| pZF-sol-R3 | P1-[mutated activator region]-lacZ fusion in pLacZF | This study |
| pZF-sol-R1/R2 | P1-[mutated activator region]-lacZ fusion in pLacZF | This study |
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen.
Analysis of fermentation products.
Samples of C. acetobutylicum DSM 792 cells were centrifuged at 20,000 × g for 10 min. One milliliter of the supernatant fluid was acidified with 0.1 ml of 2 N HCl containing 110 mM isobutanol (final concentration in the sample, 10 mM) as an internal standard. Subsequently, 1 μl was used for detection and quantification of fermentation products with a Chrompack CP9001 gas chromatograph equipped with a flame ionization detector (Chrompack GmbH, Frankfurt, Germany). The following products were analyzed: acetate, acetone, butanol, butyrate, and ethanol. Separation took place in a Chromosorb 101 column (length, 2 m; 80 to 100 mesh) at 155 to 197°C (rate of temperature increase, 9°C per min) with N2 as the carrier gas (flow rate, 30 ml per min). The injector temperature was 195°C, and the detector temperature was 230°C. Signal analysis was performed by using the program Maestro II (version 2.1).
Heterologous overexpression and purification of AdhE.
For heterologous expression of AdhE in E. coli, plasmid pTWa4-2::adhE6×His was constructed by generating His-tag-encoding adhE by PCR performed with primers KLADHE01 and KLADHE02 (Table 2) and with plasmid pK9 (9) as the template and ligating the product into vector pTWa4-2 (36) following digestion with SalI and EcoRI. All standard procedures for cloning were carried out as described elsewhere (29).
TABLE 2.
Deoxyoligonucleotides used in this study
| Primer | Sequence (5′ → 3′)a |
|---|---|
| KLADHE01 | TATTTTAGAAAGAAcTGcAgAcaTATGAAAGTCAC |
| KLADHE02 | CCTCCTTTTAAATTCTTTATgAAttcTtagtgAtGgtGaTGgtgatgaggcGTTggAGGTTGTTTTTTAAAAC |
| KL06 | GGGAATACCATAtgTcgACACTTCTTTCTAAAATA |
| KL07 | CGgAaTtCATTTAAATACACAGCTG |
| KL11 | AGATCgataTcggtAaccTCATAAAATTTATGATCC |
| KL12 | AGATCgatatcTTAAATACACAGCTG |
| KL17 | AAAAACAATATtGACATaATaTTcAAgATATAATAAAT |
| KL18 | ATTTATTATATcTTgAAtATtATGTCaATATTGTTTTT |
| AdhE-PE-nat-IRD | CTTCTTTAATTACCTTGAGT |
| AdhE-PE-klimp-IRD | GTGACTTTCATATGTCGACAC |
| 5-inter-sol-eco | GTAATTgAATTcAATGATTTAGGCATAGAAATCG |
| 5-inter-sol-pst | AATActgCAGCTGTGTATTTAAATGTAAATAGC |
| lacZ-5-Sal-frame | GGAAGGTGACTgTcgacAGAAAGATTATTCC |
| lacZ-3-Pst-end | GCTTATATAATCTGcAGATGAAATTCTC |
| adhE-fus-up-Sal | AATTCgTcgACTGTTGTGACTTTCATAAATATACAC |
| PadhE+Act-Bam | TATTGGatCcATTAATTAGGGTTATATATACTAG |
| PadhE-Act-Bam | AAAAGgatCcATTTACATTTAAATACACAGCTG |
| spo0A-up | GGAATCTAATATTTTGGaactATAATATATATTTAGG |
| spo0A-down | CCTAAATATATATTATagttCCAAAATATTAGATTCC |
| R1-up | GGAATTATCAGTAaAcTaAtAAAAATATGAAGG |
| R1-down | CCTTCATATTTTTaTtAgTtTACTGATAATTCCTAA |
| R2-up | GCGTAATAATATAaAcTaAtGAATTATCAGTATATTTAG |
| R2-down | CTAAATATACTGATAATTCaTtAgTtTATATTATTACGCC |
| R3-up | CAAAATGGTATCTAAcAagaTGGCGTAATATATATTTAGG |
| R3-down | ATATATATTACGCCAtctTgTTAGATACCATTTTGAAAAG |
| R1/2-up | CGTAATATAaAcTaAtGAATTATCAGTAaAcTaAtAAAAATATG |
| R1/2-down | CATATTTTTaTtAgTtTACGTATAATTCaTtAgTtTATATTACG |
Substituted bases are indicated by lowercase letters; introduced restriction sites are indicated by italics.
E. coli WL3(pGP1-2) transformed with pTWa4-2::adhE6×His was grown aerobically in Luria-Bertani medium. When an optical density at 600 nm of 0.7 to 0.8 was reached, expression of His6-tagged AdhE was induced by adding 1 mM isopropyl-β-thiogalactopyranoside (IPTG). After an additional 3 h of growth, the cells were harvested by centrifugation (5,000 × g, 10 min, 4°C). A crude extract was prepared by washing and suspending the cells in imidazole buffer (50 mM Na2HPO4 [pH 8.0], 300 mM NaCl, 10 mM imidazole), followed by three passages through a French press (SLM Instruments Company, Urbana, Ill.) at 12.5 MPa and centrifugation (30 min, 30,000 × g, 4°C). The His6-tagged AdhE and butanol dehydrogenase domain proteins were purified by using nickel nitriloacetic acid (Ni-NTA) agarose with the buffers recommended by the manufacturer (Qiagen GmbH, Hilden, Germany) at 4°C. Protein eluted at 125 mM imidazole. Subsequently, a second purification step was carried out by using a column packed with 2 ml of Reactive Green 19 (Sigma-Aldrich Chemie GmbH, Deisenhofen, Germany) and equilibrated with 5 bed volumes of 20 mM Tris-HCl (pH 7.5). The fractions obtained from the first purification step were loaded onto the column, incubated for 30 min, and washed with 2 bed volumes of equilibration buffer containing 200 mM NaCl. Finally, the protein was eluted from the column by increasing the concentration of NaCl in the buffer to 750 mM.
Detection of AdhE expression in C. acetobutylicum.
To construct plasmids pKLIMP7, pKLIMP12, and pKLIMP17, the sol promoter region was amplified from plasmid pK9 (9) and cloned upstream of the adhE gene into pTWa4-2::adhE6×His. The sol promoter fragment for pKLIMP7 was obtained by using primers KL06 and KL07, and the fragments for pKLIMP12 and pKLIMP17 were amplified with primer pairs KL06-KL11 and KL06-KL12, respectively (Table 2). Site-specific mutations in the sol promoter region in pKLIMP17 were created with primers KL17 and KL18 by the overlap extension-PCR technique (11). Following digestion with EcoRV and NdeI, the fragments for pKLIMP12 and pKLIMP17 (lengths, 217 and 305 bp) were cloned into pTWa4-2::adhE6×His that was cut with the same enzymes. The fragment for pKLIMP7 was digested with EcoRI and NdeI and, after treatment of the EcoRI site with polymerase I, inserted as a blunt NdeI fragment (290 bp) into the NdeI/XhoI restriction site of pTWa4-2::adhE6×His after the vector's XhoI site was trimmed with polymerase I. Finally, the sol promoter region-His6-tag-encoding adhE fusions were transferred as EcoRV/EcoRI fragments (lengths, 3.1, 2.85, and 2.9 kbp, respectively) into SmaI/EcoRI-digested vector pIMP1 (19) to obtain plasmids pKLIMP7, pKLIMP12, and pKLIMP17. The constructs were methylated in E. coli ER2275(pAN1) (20) and subsequently used to electrotransform C. acetobutylicum DSM 792 (24).
His6-tagged AdhE expression was detected with cultures grown in MES-buffered minimal medium (500 ml) after the onset of solventogenesis, indicated by formation of acetone. The cells in 250-ml portions of the cultures were harvested by centrifugation (5,000 × g, 10 min, 4°C). Crude extracts were prepared by washing the cells, suspending them in phosphate buffer (50 mM KH2PO4 [pH 7.0], 10% [vol/vol] glycerol), and disrupting them by six passages through a French press (SLM Instruments Company) at 12.5 MPa, followed by centrifugation (30 min, 30,000 × g, 4°C). Subsequently, samples were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (14) and transferred onto a nitrocellulose membrane (Hybond ECL; Amersham Buchler GmbH & Co. KG, Braunschweig, Germany) as described elsewhere (13) using a Multiphor II NovaBlot unit (Amersham Pharmacia Biotech Europe GmbH, Freiburg, Germany). Then His-tagged proteins were detected with Ni-alkaline phosphatase conjugate used as recommended by the manufacturer (Qiagen GmbH).
N-terminal sequencing was performed with an ABI 477A automated sequencer (Applied Biosystems, Foster City, Calif.) after transfer of proteins onto a polyvinylidene difluoride membrane (Millipore GmbH, Eschborn, Germany). This was done at the Department of Biochemistry II, University of Göttingen, Göttingen, Germany.
Primer extension analyses.
C. acetobutylicum DSM 792 RNA was isolated by using an RNeasy kit and a QIAshredder (Qiagen GmbH) according to the manufacturer's instructions. Primer extension analyses were carried out as described elsewhere (12) with the IRD800-labeled primers AdhE-PE-nat-IRD (native promoter) and AdhE-PE-klimp-IRD (pKLIMP17) (Table 2). The signals were analyzed with an automatic sequencer (LI-COR 4000L; Licor, Inc., Lincoln, Nebr.) using a 6% (wt/vol) polyacrylamide gel at 1,500 V and 50°C.
Cloning and directed mutagenesis of the sol activator region.
A PCR fragment (454 bp) containing the C-terminal part of orf5 and the intergenic region between orf5 and the sol promoter was amplified from plasmid pK9 (9) by using primers 5-inter-sol-eco and 5-inter-sol-pst (Table 2). After digestion with EcoRI and PstI the fragment was cloned in EcoRI/PstI-cut vector pIMP1 (19), which yielded plasmid pACT1.
The reporter vector pLacZF was constructed by cloning the lacZ gene encoding the β-galactosidase of Thermoanaerobacterium thermosulfurigenes (5) as a PCR fragment (2,318 bp) amplified with primers lacZ-5-Sal-frame and lacZ-3-Pst-end (Table 2) from plasmid pSN51 into pIMP1 after trimming with SalI and PstI.
PCR fragments containing the sol promoter region were amplified from pK9 (9) by using primers adhE-fus-up-Sal and PadhE+Act-Bam (574 bp; pZF-sol) and primers adhE-fus-up-Sal and PadhE-Act-Bam (346 bp; pZF-sol-act) (Table 2). Following digestion with SalI and BamHI the fragments were cloned into SalI/BamHI-cut pLacZF to obtain plamids pZF-sol and pZF-sol-act, which provided an adhE-lacZ fusion. Site-specific mutations were introduced by overlap extension-PCR (11) by using primers spo0A-up and spo0A-down, primers R1-up and R1-down, primers R2-up and R2-down, and primers R3-up and R3-down (Table 2) to generate plasmids pZF-sol-spo0A, pZF-sol-R1, pZF-sol-R2, and pZF-sol-R3. For the double mutant pZF-sol-R1/R2, primers R1/2-up and R1/2-down (Table 2) were used for the mutation of vector pZF-sol-R1.
In vivo analysis of the sol activator region.
Plasmids pZF-sol, pZF-sol-act, pZF-sol-spo0A, pZF-sol-R1, pZF-sol-R2, pZF-sol-R1/R2, and pZF-sol-R3 were methylated in vivo and transferred to C. acetobutylicum DSM 792 by electroporation as described above for the pKLIMP vectors. Subsequent growth experiments were carried out in 500-ml cultures of MES-buffered minimal medium. After exponential growth began, 20-ml samples were harvested at 2-h intervals. Each supernatant was used for product analysis, and the pellet was suspended in 500 μl of buffer (50 mM KH2PO4 [pH 7.0], 10% [wt/vol] glycerol) and mixed with 250 mg of glass beads (Sigma-Aldrich Chemie GmbH). The cells were subsequently disrupted with a Ribolyser cell disrupter (Hybaid GmbH, Heidelberg, Germany) at maximal intensity three times (45 s each). The sample was centrifuged (20,000 × g, 30 min, 4°C), and the supernatant was immediately processed for β-galactosidase assays. The assays were carried out using an Ultrospec3000 spectrophotometer (Amersham Pharmacia Biotech GmbH, Freiburg, Germany). An 840-μl portion of phosphate buffer (50 mM KH2PO4, pH 7.0) and 40 μl of o-nitro-β-d-galactopyranoside (4 mg/ml) were mixed and kept at 65°C for 5 to 10 min. The reaction was started by adding 20 μl of crude extract. Activity was indicated by a linear increase in extinction due to the conversion of o-nitro-β-d-galactopyranoside. A specific coefficient of extinction for o-nitro-β-d-galactopyranoside of 3.5 mM−1·cm−1 was used.
Protein determination was carried out by the method of Bradford (4). Bio-Rad protein assay solution (Bio-Rad Laboratories, Munich, Germany) was used as the dye reagent, and bovine serum albumin or ovalbumin was used as the standard protein.
RESULTS
Analysis of the sol operon promoter region.
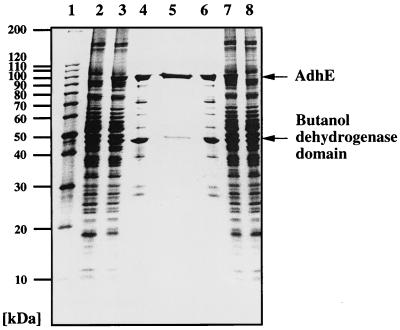
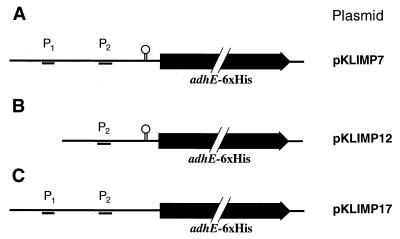
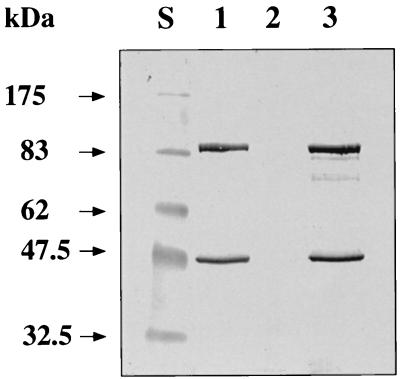
During investigations aimed at purification and characterization of the AdhE protein, we constructed a plasmid carrying the adhE gene with a 3′-fused tail encoding six histidines under control of a T7 promoter (pTWa4-2::adhE6×His). The plasmid was purified by successive affinity chromatography on Ni-NTA and affinity chromatography on Reactive Green 19 after heterologous expression in E. coli. Surprisingly, two proteins with apparent molecular masses of 98 and 48 kDa were copurified (Fig. 1). The size of the 98-kDa protein corresponds nicely to the expected size of AdhE (96 kDa) plus an additional His tag and an immunoglobulin A protease cutting site. The identity of this protein was confirmed by N-terminal amino acid sequencing. The first 10 amino acids (Met-Lys-Val-Thr-Thr-Val-Lys-Glu-Leu-Asp) matched exactly the beginning of AdhE, as deduced from the nucleotide sequence (9). The second protein started with the sequence Met-Leu-?-Phe-Arg-Val-Pro-?-Lys-Val-Tyr-Phe-Lys-Phe-Gly (question marks represent residues not unambiguously identified during sequencing). This sequence corresponds to the beginning of the second domain of AdhE, the alcohol dehydrogenase domain (amino acids 449 to 463) (9). A ribosome binding site (5′-AGGAGA-3′) is separated by 6 bp from an ATG codon encoding methionine, thus explaining the second translation start from the same mRNA in E. coli. The size of the 48-kDa protein corresponds nicely to the expected size of that domain (46 kDa) plus an additional His tag and an immunoglobulin A protease cutting site. To check whether this result was an artifact resulting from expression in a heterologous host or whether it mimicked the natural conditions, we constructed two shuttle vectors carrying the adhE gene with a 3′-fused tail encoding six histidines and (i) the complete promoter region (pKLIMP7) or (ii) only putative proximal promoter P2 (pKLIMP12) (Fig. 2). After transformation into C. acetobutylicum and preparation of protein extracts, Western blotting revealed that the two translational start sites were also used in the natural host (Fig. 3). However, a surprising result was that no AdhE was synthesized from the plasmid with only putative proximal promoter P2. This finding suggests that the sol operon of C. acetobutylicum is controlled by only a single promoter, namely P1 (or S2).
FIG. 1.
Purification of clostridial mature AdhE and the butanol dehydrogenase domain from the heterologous host E. coli. Separation was performed with an SDS-6 to 25% (wt/vol) polyacrylamide linear gradient gel and was followed by silver staining. Lane 1, marker proteins; lanes 2 and 8, crude extract from WL3(pGP1-2, pTWa4-2) (negative control); lanes 3 and 7, crude extract from WL3(pGP1-2, pTWa4-2::adhE6×His); lanes 4 and 6, fractions after affinity chromatography with Ni-NTA; lane 5, fraction after affinity chromatography with Reactive Green 19.
FIG. 2.
Plasmids constructed with targeted mutations in the sol operon regulatory region of C. acetobutylicum. Only the regulatory region and the first structural gene (adhE, altered by a six-histidine-encoding tail) are shown; these regions were inserted into the E. coli-C. acetobutylicum shuttle vector pIMP1. P1 and P2 are putative promoter structures deduced from primer extension studies. The hairpin symbol indicates a stem-loop structure predicted by the computer program MFold. For further details see the text.
FIG. 3.
Colorimetric detection of His-tagged AdhE and butanol dehydrogenase domain in recombinant C. acetobutylicum strains. Cell extracts were separated by SDS-polyacrylamide gel electrophoresis and were transferred onto a nitrocellulose membrane. Subsequently, His-tagged proteins were detected with Ni-AP conjugate. The molecular sizes of marker proteins are indicated on the left. Lane S, marker proteins; lane 1, C. acetobutylicum(pKLIMP7): lane 2, C. acetobutylicum(pKLIMP12); lane 3, C. acetobutylicum(pKLIMP17).
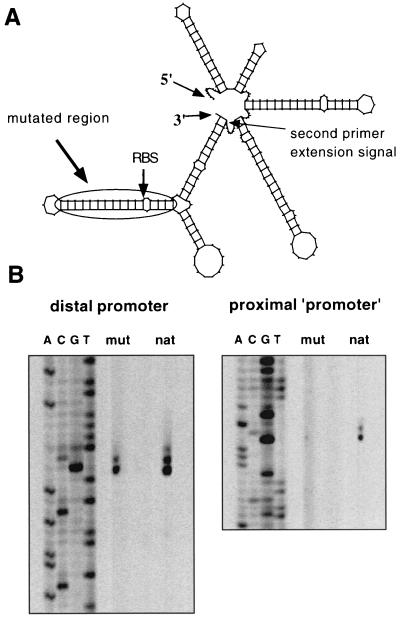
Computer programs (MFold and PlotFold [38]) indicated that the 246-base noncoding region between P1 and the ATG start codon of AdhE folds into a very complex secondary structure (Fig. 4A). Several stem-loop structures were predicted; however, only the base of one structure yielded the second primer extension signal (P2). This argues against the hypothesis that there is a nonspecific, artificial falling off of the reverse transcriptase during the primer extension reaction. To verify that only one promoter controls the sol operon, another plasmid was constructed. pKLIMP17 was identical to pKLIMP7 except that a stem-loop structure directly upstream of adhE was disrupted by targeted PCR mutagenesis (Fig. 2). Specific primer extension experiments with the recombinant C. acetobutylicum strain could be performed by using a primer complementary to the artificially introduced SalI and NdeI cutting sites upstream of the adhE structural gene. These sites had been introduced for plasmid construction (see Materials and Methods). In the wild type, two signals (corresponding to the P1 and P2 start points) were detected, as expected. However, pKLIMP17 yielded only the transcription start site originating from P1 (Fig. 4B). Due to the distance between the stem-loop mutations and P2, these mutations are unlikely to affect any putative promoter regulation. Thus, only P1 controls expression of the sol operon. The finding that in pKLIMP17 no P2 signal could be detected can be explained by the altered secondary structure of the whole region due to disruption of the stem-loop structures adjacent to the beginning of adhE. This suggests that there is specific mRNA processing (with an RNase recognizing the specific secondary structure) rather than a nonspecific falling off of the reverse transcriptase during cDNA formation.
FIG. 4.
Secondary structure of the 5′ untranslated region of the sol operon transcript and effect of targeted mutations on transcription start mapping by primer extension. (A) Secondary structure of the beginning of the sol operon transcript, as determined by the program MFold and plotted by the program PlotFold. The positions of the second primer extension signal, which led to deduction of P2, and the region used for targeted mutagenesis are indicated. RBS, ribosome-binding site. (B) Primer extension experiments showing signals related to P1 (distal promoter) and P2 (proximal start site). The results of sequence reactions, which were obtained by using the same oligodeoxynucleotides, are shown on the left in each gel. mut, mutated region; nat, natural sequence.
Identification of a regulator-binding region of the sol operon.
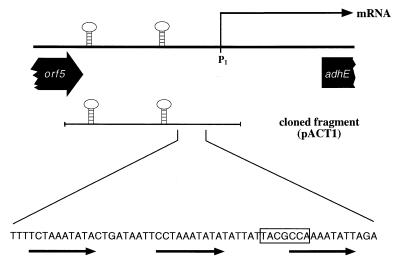
Recent experiments have provided evidence that purified Orf5 protein does not bind to the regulatory region of the sol operon, either in the form of a linear template or in a supercoiled plasmid (33). Although all of the data obtained for purified Orf5 indicated that it is involved in glycosylation or deglycosylation and that it does not repress solventogenesis, the reported result of its overproduction (i.e., inhibition of solvent formation) (22) could not be reproduced and remained unexplained (33). A detailed examination of the subcloning procedures used in the previous study revealed that the insert used for overexpression of Orf5 also contained part of the intergenic region of the adjacent sol operon, directly upstream of the P1 promoter. Especially intriguing in this area was a motif consisting of three incomplete repeats (10 bp each), which were separated by 9 or 10 bp (Fig. 5). The second repeat (repeat 2 [R2]) had the sequence 5′-CCTAAATATA-3′, and there were just one or two mismatches in R1 and R3 at the first or third and last nucleotides, respectively. R3 also partially overlapped with a putative Spo0A-binding motif, which was proposed recently (28). The lengths and spacing of these repeats would allow contact by a regulatory protein from one side of the DNA. Also, the location of this motif in the region 50 to 110 bp upstream of the sol operon promoter P1 (the only transcriptional control site, as shown above) would be perfectly suited for interaction of a transcription factor with RNA polymerase. Therefore, a 430-bp fragment carrying the intergenic region between orf5 and the P1 promoter was subcloned in shuttle plasmid pIMP1, yielding plasmid pACT1, and then transformed into C. acetobutylicum. After batch cultivation of the recombinant and a control strain carrying only the shuttle vector, the patterns of fermentation products were compared (Table 3). The results correspond nicely to the data reported by Nair et al. (22) for overexpression of Orf5; virtually no acetone or butanol was formed, the amount of ethanol was decreased, and butyrate production was significantly increased compared to the control strain. Thus, the solvent-negative phenotype is not caused by overexpression of the orf5 gene product (acting as a repressor) but instead is caused by the presence of several copies of a short fragment of DNA carrying a regulatory locus. This finding suggests that there is an activator for sol operon regulation, which is titrated out by multiple copies of the regulatory region.
FIG. 5.
Regulatory motifs in the sol operon regulatory region. The top line schematically shows the end of the orf5 gene with its two transcriptional terminators (hairpins), the position of the P1 promoter as the start point of the mRNA, and the beginning of the adhE gene. The second line indicates the fragment used to construct pACT1. The sequence at the bottom is the sequence of the DNA region comprising the three incomplete repeats (from left to right, R1, R2, and R3) and the Spo0A-binding site (box).
TABLE 3.
Product formation in recombinant C. acetobutylicum strains
| Plasmid | Concn of fermentation products (mM)
|
||||
|---|---|---|---|---|---|
| Solvents
|
Acids
|
||||
| Ethanol | Butanol | Acetone | Acetate | Butyrate | |
| pIMP1 (control) | 31.5 | 61.0 | 9.3 | 48.5 | 60.5 |
| pACT1 | 21.2 | 1.7 | 0 | 19.3 | 71.3 |
Reporter gene analyses.
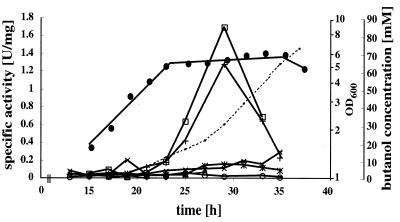
Use of the lacZ gene from T. thermosulfurigenes EM1 (5) as a reporter in C. acetobutylicum has been described previously (8, 34). For in vivo analysis of the sol operon regulatory region, we constructed a number of plasmids with targeted mutations in the three repeats and the Spo0A-binding site, followed by the P1 promoter and the lacZ gene, as shown in Fig. 6. Since R3 and the Spo0A-binding site partially overlapped, mutagenesis affected only the individual nucleotides. After in vivo methylation and electroporation into C. acetobutylicum, the recombinant strains were grown in MES-buffered minimal medium and monitored at regular intervals for β-galactosidase activity (Fig. 7). Synthesis of the enzyme paralleled induction of butanol formation. The peak pattern indicated that there was rapid degradation of the heterologous β-galactosidase in the stationary growth phase, which is consistent with stability determinations reported previously (34). Disruption of R1, R3, and the putative Spo0A-binding motif completely eliminated the reporter activity. Mutagenesis of R2, however, had no effect. Expression of β-galactosidase was prevented only in combination with disrupted R1. These results confirm the importance of this region for regulation of the sol operon. The putative Spo0A-binding motif was proposed previously based on sequence homology (28), and shutdown of reporter activity upon disruption of this sequence indicates that this transcription factor is indeed a central element in sol operon control.
FIG. 6.
Targeted mutations in the sol operon regulatory region. The top sequence is the wild-type nucleotide sequence in the reporter gene plasmid pZF-sol; the positions of the three incomplete repeats and the Spo0A-binding site (shaded box) are indicated. The other sequences indicate the mutations in the various motifs (exchanged nucleotides are indicated by boldface type).
FIG. 7.
LacZ expression from wild-type and mutant regulatory regions of the sol operon in vivo. Growth (•), butanol formation (dotted line), and β-galactosidase activity were assayed during the bacterial growth cycle for strains carrying the plasmids shown in Fig. 6. Symbols: □, wild type (pZF-sol); ◊, pZF-sol-R1; +, pZF-sol-R2; ×, pZF-sol-R3; ○, pZF-solR1/R2; ∗, pZF-sol-spo0A; ▵, pZF-sol-act. Growth and butanol formation patterns were virtually identical in all experiments. For the sake of clarity, only the data obtained with pZF-solR1/R2 are shown.
DISCUSSION
The results reported here provide conclusive evidence that the sol operon of C. acetobutylicum is transcribed only from the distal start point P1. Our data are in agreement with the almost perfect homology of the deduced promoter sequence and the consensus sequence of σA-controlled promoters. The second signal observed most likely resulted from processing of the primary transcript, which takes place in C. acetobutylicum but not in the heterologous host E. coli (9). Expression of the sol operon thus closely resembles transcription of the E. coli adhE gene, which encodes a multifunctional acetaldehyde/ethanol dehydrogenase (plus a regulatory function) and exhibits extensive sequence homology with the adhE gene of C. acetobutylicum (9, 10). In E. coli, two start points of the adhE gene transcript have also been detected (292 and 188 bp upstream of the ATG codon), and only the distal one exhibited sequence homology with a consensus promoter (1). In agreement with the situation found in C. acetobutylicum, the signal stemming from the proximal point was much more intense. The investigations of E. coli revealed that the presence of RNase III was required for successful expression of the adhE gene, suggesting that the second (proximal) signal is a result of mRNA processing (1). Although later investigations showed that the region upstream of the proximal start point could act as a promoter in transcriptional and translational lacZ fusions, it is not yet known under what natural conditions this promoter becomes active (18). In C. acetobutylicum a second adhE gene is present on the megaplasmid; this gene is induced only when reduced substrates, such as glycerol, are provided and results in only butanol and ethanol formation (L. Fontaine, I. Meynial-Salles, and P. Soucaille, Clostridium 2000, 6th Int. Workshop Regul. Metab. Genet. Dev. Solvent Acid Forming Clostridia, poster presentations, 2000), and it is also preceded by a large untranslated DNA region, as determined by DNA sequence analysis. No data concerning regulation of this gene are available yet. However, not all genes homologous to adhE are controlled in a similar manner. In Lactococcus lactis, no large untranslated region is present upstream of the start codon, and only a single transcriptional start site has been identified (2). It is interesting that in E. coli the adhE transcript is obviously processed by RNase III (1). So far, no detailed investigations of RNases in C. acetobutylicum have been described. However, transcript processing in this organism has been observed with the gap operon message, and sequence comparisons indicated that there are RNase III and RNase E-like enzymes in C. acetobutylicum (31). Whether the endonucleolytic cleavage of the sol operon transcript stabilizes the message or plays an additional regulatory role is not known at this point.
An interesting phenomenon is the second translation start point within the adhE gene, which leads to separate expression of the butanol dehydrogenase domain. The alternative, cleavage of the mature AdhE into two proteins, seems unlikely, since during protein purification no band at a size corresponding to the size of the butyraldehyde dehydrogenase domain was observed. The physiological relevance of this start point is not known yet. It might be a means to ensure a certain level of enzyme activity in the cell in case the large bifunctional protein is more unstable. So far, attempts to detect enzymatic activity in purified AdhE preparations have failed, which might support this hypothesis. However, detailed analyses of the purified butanol dehydrogenase domain, as well as the complete enzyme, are required to answer this question unambiguously.
Identification of a regulatory region upstream of P1 provides an explanation for the solvent-negative phenotype of orf5-overexpressing strains reported by Nair et al. (22). The construct used in the study of Nair et al. carried in addition to the orf5 gene the three-repeat motif and the Spo0A-binding motif. Thus, it is now clear that the sol operon is not regulated by a putative repressor but rather is regulated through transcriptional activation, most probably by Spo0A (as demonstrated by the lack of reporter activity upon disruption of the Spo0A-binding motif). This conclusion is in perfect agreement with the recent report that the other operon essential for solvent formation, the monocistronic adc gene, is also controlled by Spo0A (28). Of the three repeats, only R1 and R3 were found to be essential for regulation. While R3 partially overlaps with the putative Spo0A-binding motif and the observed effect might be caused by Spo0A binding as well, R1 is located much farther upstream and thus is unlikely to be affected by Spo0A binding. This indicates that an additional activator protein, which acts in concert with Spo0A, is involved. This hypothesis is supported by the results obtained after only the regulatory region (solvent-negative phenotype) was subcloned. Such a phenomenon is typical of titrating out a transcription factor by providing multiple binding sites. The plasmid used has a copy number of six to eight (15). However, it is unlikely that Spo0A could be titrated out by such a low number of binding sites, as numerous similar motifs have been detected in C. acetobutylicum (28). Thus, the data indicate that there is an additional activator, which may interact with Spo0A. This putative activator should be specific for the sol operon, since no other R1-like motifs could be detected upstream of adc and the second adhE gene (or in the whole genome, which has recently been described [25]). At first glance, it might be surprising that transformation of only the regulatory region caused a solvent-negative phenotype, while the same region in the reporter gene constructs did not affect butanol formation. We think that this effect is caused by the presence of the P1 promoter in the latter plasmids. Initiation of transcription by RNA polymerase causes increased negative supercoiling upstream of the promoter, according to the twin transcriptional loop model (16). This event might result in a falling off of the activator, which in turn is available for binding at the same site in another plasmid. Thus, even three or four copies of the second activator protein would be sufficient to ensure onset of butanol formation in the presence of several plasmids with the same binding motif. Support for this hypothesis comes from data showing that prevention of negative supercoiling leads to a dramatic increase in transcription of the sol operon (35) (anaerobic expression of adhE in E. coli is also influenced by DNA topology [18]) and that under natural conditions the onset of solventogenesis in C. acetobutylicum is paralleled by relaxation of DNA (37). Thus, only the presence of multiple binding sites without the possibility of changing the degree of DNA supercoiling would result in irreversible binding of the activator and prevention of transcription initiation at the proper sol operon regulatory site (as observed in our experiments). In future experiments we will try to identify and characterize the additional activator protein.
Acknowledgments
We thank Bernhard Schmidt for N-terminal protein sequencing. Clarithromycin was a generous gift from Abbott Laboratories, Queenborough, United Kingdom.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Aristarkhov, A., A. Mikulskis, J. G. Belasco, and E. C. C. Lin. 1996. Translation of the adhE transcript to produce ethanol dehydrogenase requires RNase III cleavage in Escherichia coli. J. Bacteriol. 178:4327-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnau, J., F. Jørgensen, S. M. Madsen, A. Vrang, and H. Israelsen. 1998. Cloning of the Lactococcus lactis adhE gene, encoding a multifunctional alcohol dehydrogenase, by complementation of a fermentative mutant of Escherichia coli. J. Bacteriol. 180:3049-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertram, J., A. Kuhn, and P. Dürre. 1990. Tn916-induced mutants of Clostridium acetobutylicum defective in regulation of solvent formation. Arch. Microbiol. 153:373-377. [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Burchhardt, G., and H. Bahl. 1991. Cloning and analysis of the β-galactosidase-encoding gene from Clostridium thermosulfurogenes EM1. Gene 106:13-19. [DOI] [PubMed] [Google Scholar]
- 6.Cornillot, E., and P. Soucaille. 1996. Solvent-forming genes in clostridia. Nature 380:489. [Google Scholar]
- 7.Cornillot, E., R. Nair, E. T. Papoutsakis, and P. Soucaille. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J. Bacteriol. 179:5442-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dürre, P., R.-J. Fischer, A. Kuhn, K. Lorenz, W. Schreiber, B. Stürzenhofecker, S. Ullmann, K. Winzer, and U. Sauer. 1995. Solventogenic enzymes of Clostridium acetobutylicum: catalytic properties, genetic organization, and transcriptional regulation. FEMS Microbiol. Rev. 17:251-262. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, R.-J., J. Helms, and P. Dürre. 1993. Cloning, sequencing, and molecular analysis of the sol operon of Clostridium acetobutylicum, a chromosomal locus involved in solventogenesis. J. Bacteriol. 175:6959-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodlove, P. E., P. R. Cunningham, J. Parker, and D. P. Clark. 1989. Cloning and sequence analysis of the fermentative alcohol-dehydrogenase-encoding gene of Escherichia coli. Gene 85:209-214. [DOI] [PubMed] [Google Scholar]
- 11.Ho, S. N., H. D. Hunt, R. M. Morton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 12.Kellmann, J.-W., E. Piechersky, and B. Piechulla. 1990. Analysis of the diurnal expression patterns of the tomato chlorophyll a/b binding protein genes. Influence of light and characterization of the gene family. Phytochem. Phytobiol. 52:35-41. [DOI] [PubMed] [Google Scholar]
- 13.Khyse-Andersen, J. 1984. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10:203-209. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. Y., G. N. Bennett, and E. T. Papoutsakis. 1993. Determination of plasmid copy number and stability in Clostridium acetobutylicum ATCC 824. FEMS Microbiol. Lett. 108:319-324. [DOI] [PubMed] [Google Scholar]
- 16.Liu, L. F., and J. C. Wang. 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 84:7024-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorowitz, W., and D. P. Clark. 1982. Escherichia coli mutants with a temperature-sensitive alcohol dehydrogenase. J. Bacteriol. 152:935-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Membrillo-Hernández, J., and E. C. C. Lin. 1999. Regulation of expression of the adhE gene, encoding ethanol oxidoreductase in Escherichia coli: transcription from a downstream promoter and regulation by Fnr and RpoS. J. Bacteriol. 181:7571-7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mermelstein, L. D., N. E. Welker, G. N. Bennett, and E. T. Papoutsakis. 1992. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Bio/Technology 10:190-195. [DOI] [PubMed] [Google Scholar]
- 20.Mermelstein, L. D., and E. T. Papoutsakis. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage Φ3TI to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 59:1077-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair, R. V., G. N. Bennett, and E. T. Papoutsakis. 1994. Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. J. Bacteriol. 176:871-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nair, R. V., E. M. Green, D. E. Watson, G. N. Bennett, and E. T. Papoutsakis. 1999. Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor. J. Bacteriol. 181:319-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakotte, S. 1998. Funktionelle Analyse von Regulationsstrukturen essentieller Gene für die Lösungsmittelbildung in Clostridium acetobutylicum. Ph.D. thesis. University of Ulm, Ulm, Germany.
- 24.Nakotte, S., S. Schaffer, M. Böhringer, and P. Dürre. 1998. Electroporation of, plasmid isolation from, and plasmid conservation in Clostridium acetobutylicum DSM 792. Appl. Microbiol. Biotechnol. 50:564-567. [DOI] [PubMed] [Google Scholar]
- 25.Nölling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, GTC Sequencing Center Production, Finishing, and Bioinformatics Teams, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oultram, J. D., M. Loughlin, T.-J. Swinfeld, J. K. Brehm, D. E. Thomson, and N. P. Minton. 1988. Introduction of plasmids into whole cells of Clostridium acetobutylicum by electroporation. FEMS Microbiol. Lett. 56:83-88. [Google Scholar]
- 27.Petersen, D. J., R. W. Welch, F. B. Rudolph, and G. N. Bennett. 1991. Molecular cloning of an alcohol (butanol) dehydrogenase gene cluster from Clostridium acetobutylicum ATCC 824. J. Bacteriol. 173:1831-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravagnani, A., K. C. B. Jennert, E. Steiner, R. Grünberg, J. R. Jeffries, S. R. Wilkinson, D. I. Young, E. C. Tidswell, D. P. Brown, P. Youngman, J. G. Morris, and M. Young. 2000. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol. Microbiol. 37:1172-1185. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sauer, U., and P. Dürre. 1995. Differential induction of genes related to solvent formation during the shift from acidogenesis to solventogenesis in continuous culture of Clostridium acetobutylicum. FEMS Microbiol. Lett. 125:115-120. [Google Scholar]
- 31.Schreiber, W., and P. Dürre. 2000. Differential expression of genes within the gap operon of Clostridium acetobutylicum. Anaerobe 6:291-297. [Google Scholar]
- 32.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thormann, K., and P. Dürre. 2001. Orf5/SolR: a transcriptional repressor of the sol operon of Clostridium acetobutylicum? J. Ind. Microbiol. Biotechnol. 27:307-313. [DOI] [PubMed] [Google Scholar]
- 34.Tummala, S. B., N. E. Welker, and E. T. Papoutsakis. 1999. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ullmann, S., A. Kuhn, and P. Dürre. 1996. DNA topology and gene expression in Clostridium acetobutylicum: implications for the regulation of solventogenesis. Biotechnol. Lett. 18:1413-1418. [Google Scholar]
- 36.Wagner, T. 1993. Entwicklung eines Verfahrens zur Proteinproduktion in Escherichia coli unter Ausnutzung der Phasduktion am Beispiel der DCM-DNA-Cytosin-(C5)-Methyltransferase aus Escherichia coli K12. Ph.D. thesis. University of Göttingen, Göttingen, Germany.
- 37.Wong, J., and G. N. Bennett. 1996. The effect of novobiocin on solvent production by Clostridium acetobutylicum. J. Ind. Microbiol. 16:354-359. [DOI] [PubMed] [Google Scholar]
- 38.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]