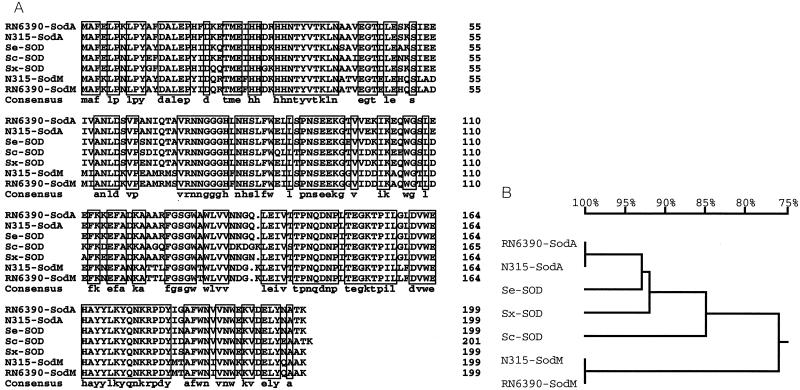
FIG. 4.
(A) Amino acid sequence alignments of staphylococcal SODs. The S. aureus RN6390 SodA (48) and SodM (48) (AF273269) proteins, the S. aureus N315 SodA and SodM proteins (36) (AP003129 and AP003134, respectively), and the Sod proteins of S. epidermidis (Se) ATCC 12228 (AF410177), S. carnosus (Sc) (CAC14833), and S. xylosus (Sx) (CAB95744) are shown. Consensus sequences are boxed. A gap at position 136 of the SodM sequences was manually inserted. (B) Phylogenetic tree. Amino acid sequences were analyzed with DNAMAN (Lynnon BioSoft, Vaudreuil, Quebec, Canada) software, which uses the neighbor-joining method described by Saitou and Nei (43).