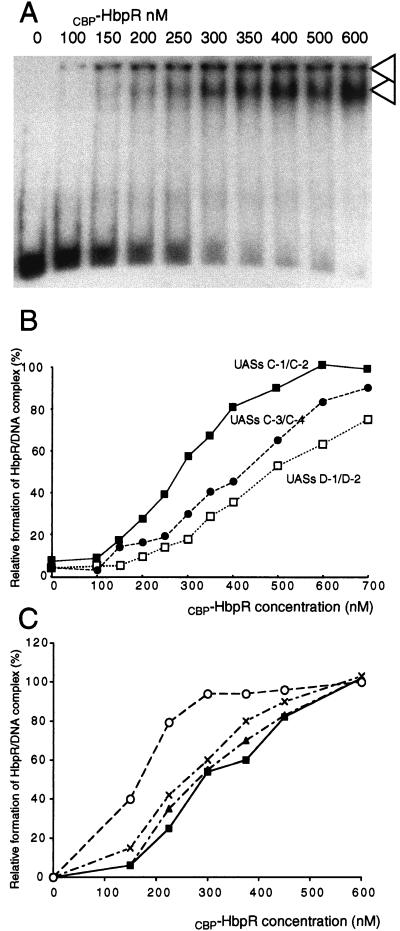
FIG. 6.
Affinity characteristics of CBP-HbpR. (A) GMSA of fragment hbpCCC6 containing UASs C-1 and C-2 with increasing concentrations of CBP-HbpR (indicated above lanes). The amount of radiolabeled DNA fragment in the assay corresponded to 80 fmol. The arrowheads point to complex 1 (top) and complex 2 (bottom) (see text). (B) Graphic representation of the relative densities of the CBP-HbpR-DNA complexes formed with increasing CBP-HbpR concentrations with fragments containing UASs C-1 and C-2, UASs C-3 and C-4, and UASs D-1 and D-2. Relative densities were calculated by laser scanning densitometric analysis of band darkness on autoradiograms (as shown in panel A) and represent the darkness of the protein-DNA complex in each lane compared to that in the incubation with 600 nM HbpR. (C) Graphic representation of the relative densities of the CBP-HbpR-DNA complexes formed with increasing CBP-HbpR concentrations with fragments containing UASs C-1 and C-2 in the presence of ATP (multiplication signs), 2-HBP (triangles), and ATP and 2-HBP (open circles) or in the absence of an inducer (closed squares). ATP was used at a concentration of 5 mM, and 2-HBP was used at 10 μM. Relative densities were calculated as explained for panel B.